Pre-ciliated tubal epithelial cells are prone to initiation of high-grade serous ovarian carcinoma
- PMID: 39366996
- PMCID: PMC11452611
- DOI: 10.1038/s41467-024-52984-1
Pre-ciliated tubal epithelial cells are prone to initiation of high-grade serous ovarian carcinoma
Erratum in
-
Author Correction: Pre-ciliated tubal epithelial cells are prone to initiation of high-grade serous ovarian carcinoma.Nat Commun. 2024 Nov 20;15(1):10053. doi: 10.1038/s41467-024-54465-x. Nat Commun. 2024. PMID: 39567528 Free PMC article. No abstract available.
Abstract
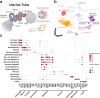
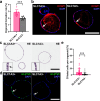
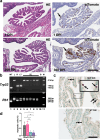
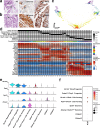
The distal region of the uterine (Fallopian) tube is commonly associated with high-grade serous carcinoma (HGSC), the predominant and most aggressive form of ovarian or extra-uterine cancer. Specific cell states and lineage dynamics of the adult tubal epithelium (TE) remain insufficiently understood, hindering efforts to determine the cell of origin for HGSC. Here, we report a comprehensive census of cell types and states of the mouse uterine tube. We show that distal TE cells expressing the stem/progenitor cell marker Slc1a3 can differentiate into both secretory (Ovgp1+) and ciliated (Fam183b+) cells. Inactivation of Trp53 and Rb1, whose pathways are commonly altered in HGSC, leads to elimination of targeted Slc1a3+ cells by apoptosis, thereby preventing their malignant transformation. In contrast, pre-ciliated cells (Krt5+, Prom1+, Trp73+) remain cancer-prone and give rise to serous tubal intraepithelial carcinomas and overt HGSC. These findings identify transitional pre-ciliated cells as a cancer-prone cell state and point to pre-ciliation mechanisms as diagnostic and therapeutic targets.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
[Significance and expression of PAX8, PAX2, p53 and RAS in ovary and fallopian tubes to origin of ovarian high grade serous carcinoma].Zhonghua Fu Chan Ke Za Zhi. 2017 Oct 25;52(10):687-696. doi: 10.3760/cma.j.issn.0529-567X.2017.10.008. Zhonghua Fu Chan Ke Za Zhi. 2017. PMID: 29060967 Chinese.
-
[Morphologic changes of fallopian tubal epithelium in ovarian serous tumors].Zhonghua Bing Li Xue Za Zhi. 2012 Jul;41(7):433-7. doi: 10.3760/cma.j.issn.0529-5807.2012.07.001. Zhonghua Bing Li Xue Za Zhi. 2012. PMID: 22932451 Chinese.
-
Molecular requirements for transformation of fallopian tube epithelial cells into serous carcinoma.Neoplasia. 2011 Oct;13(10):899-911. doi: 10.1593/neo.11138. Neoplasia. 2011. PMID: 22028616 Free PMC article.
-
Natural history of ovarian high-grade serous carcinoma from time effects of ovulation inhibition and progesterone clearance of p53-defective lesions.Mod Pathol. 2020 Jan;33(1):29-37. doi: 10.1038/s41379-019-0370-1. Epub 2019 Sep 26. Mod Pathol. 2020. PMID: 31558785 Review.
-
Cellular models of development of ovarian high-grade serous carcinoma: A review of cell of origin and mechanisms of carcinogenesis.Cell Prolif. 2021 May;54(5):e13029. doi: 10.1111/cpr.13029. Epub 2021 Mar 25. Cell Prolif. 2021. PMID: 33768671 Free PMC article. Review.
Cited by
-
Foxn3 is required to suppress aberrant ciliogenesis in nonphotoreceptor retinal neurons.Proc Natl Acad Sci U S A. 2025 Jul 22;122(29):e2500871122. doi: 10.1073/pnas.2500871122. Epub 2025 Jul 15. Proc Natl Acad Sci U S A. 2025. PMID: 40663603
-
Aging-Associated Vacuolation of Multi-Ciliated Cells in the Distal Mouse Oviduct Reflects Unique Cell Identity and Luminal Microenvironment.Aging Cell. 2025 Jul;24(7):e70051. doi: 10.1111/acel.70051. Epub 2025 May 1. Aging Cell. 2025. PMID: 40310729 Free PMC article.
-
Multi-modal characterization of transcriptional programs that drive metastatic cascades to solid sites and ascites in ovarian cancer.bioRxiv [Preprint]. 2025 Aug 27:2025.08.26.672372. doi: 10.1101/2025.08.26.672372. bioRxiv. 2025. PMID: 40909508 Free PMC article. Preprint.
-
Aggressive Serous Carcinomas of the Female Reproductive Tract: Cancer-Prone Cell States and Genetic Drivers.Cancers (Basel). 2025 Feb 11;17(4):604. doi: 10.3390/cancers17040604. Cancers (Basel). 2025. PMID: 40002199 Free PMC article. Review.
-
Therapeutic Target Discovery for Multiple Myeloma: Identifying Druggable Genes via Mendelian Randomization.Biomedicines. 2025 Apr 5;13(4):885. doi: 10.3390/biomedicines13040885. Biomedicines. 2025. PMID: 40299486 Free PMC article.
References
-
- Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin.74, 12–49 (2024). - PubMed
-
- Seidman, J. D., Ronnett, B. M., Shih Ie, M., Cho, K. R. & Kurman, R. J. in Blaustein’s Pathology of the Female Genital Tract (eds Kurman, R. J., Ellenson, L. H. & Ronnett, B. M.) 841–966 (Springer, 2019).
-
- Carlson, J. W., Gilks, C. B. & Soslow, R. A. Tumors of the Ovary and Fallopian Tube. Vol. 16 (American Registry of Pathology, 2023).
-
- Landen, C. N. Jr., Birrer, M. J. & Sood, A. K. Early events in the pathogenesis of epithelial ovarian cancer. J. Clin. Oncol.26, 995–1005 (2008). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Dryad/10.5061/dryad.4mw6m90hm
Grants and funding
- CA260115/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA182413/CA/NCI NIH HHS/United States
- 327516/Ovarian Cancer Research Fund (Ovarian Cancer Research Fund, Inc.)
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 CA248524/CA/NCI NIH HHS/United States
- R01 CA260115/CA/NCI NIH HHS/United States
- S10 RR025502/RR/NCRR NIH HHS/United States
- DGE-2139899/NSF | Directorate for Education & Human Resources | Division of Graduate Education (DGE)
- CA182413/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- CA248524/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

