Distinctive CD39+CD9+ lung interstitial macrophages suppress IL-23/Th17-mediated neutrophilic asthma by inhibiting NETosis
- PMID: 39366998
- PMCID: PMC11452667
- DOI: 10.1038/s41467-024-53038-2
Distinctive CD39+CD9+ lung interstitial macrophages suppress IL-23/Th17-mediated neutrophilic asthma by inhibiting NETosis
Abstract
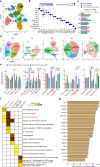
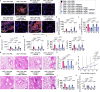
The IL-23-Th17 axis is responsible for neutrophilic inflammation in various inflammatory diseases. Here, we discover a potential pathway to inhibit neutrophilic asthma. In our neutrophil-dominant asthma (NDA) model, single-cell RNA-seq analysis identifies a subpopulation of CD39+CD9+ interstitial macrophages (IMs) suppressed by IL-23 in NDA conditions but increased by an IL-23 inhibitor αIL-23p19. Adoptively transferred CD39+CD9+ IMs suppress neutrophil extracellular trap formation (NETosis), a representative phenotype of NDA, and also Th17 cell activation and neutrophilic inflammation. CD39+CD9+ IMs first attach to neutrophils in a CD9-dependent manner, and then remove ATP near neutrophils that contribute to NETosis in a CD39-dependent manner. Transcriptomic data from asthmatic patients finally show decreased CD39+CD9+ IMs in severe asthma than mild/moderate asthma. Our results suggest that CD39+CD9+ IMs function as a potent negative regulator of neutrophilic inflammation by suppressing NETosis in the IL-23-Th17 axis and can thus serve as a potential therapeutic target for IL-23-Th17-mediated neutrophilic asthma.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

