Effects of iron supplements and iron-containing micronutrient powders on the gut microbiome in Bangladeshi infants: a randomized controlled trial
- PMID: 39367018
- PMCID: PMC11452624
- DOI: 10.1038/s41467-024-53013-x
Effects of iron supplements and iron-containing micronutrient powders on the gut microbiome in Bangladeshi infants: a randomized controlled trial
Abstract
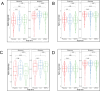
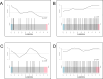
Anemia is highly prevalent globally, especially in young children in low-income countries, where it often overlaps with a high burden of diarrheal disease. Distribution of iron interventions (as supplements or iron-containing multiple micronutrient powders, MNPs) is a key anemia reduction strategy. Small studies in Africa indicate iron may reprofile the gut microbiome towards pathogenic species. We seek to evaluate the safety of iron and MNPs based on their effects on diversity, composition, and function of the gut microbiome in children in rural Bangladesh as part of a large placebo-controlled randomized controlled trial of iron or MNPs given for 3 months (ACTRN12617000660381). In 923 infants, we evaluate the microbiome before, immediately following, and nine months after interventions, using 16S rRNA gene sequencing and shotgun metagenomics in a subset. We identify no increase in diarrhea with either treatment. In our primary analysis, neither iron nor MNPs alter gut microbiome diversity or composition. However, when not adjusting for multiple comparisons, compared to placebo, children receiving iron and MNPs exhibit reductions in commensal species (e.g., Bifidobacterium, Lactobacillus) and increases in potential pathogens, including Clostridium. These increases are most evident in children with baseline iron repletion and are further supported by trend-based statistical analyses.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- World Health Organization. The global prevalence of anaemia in 2011 (World Health Organization, Geneva, 2015).
-
- World Health Organization. Global Health Observatory data repository: Anaemia in children estimates by country and WHO region (World Health Organization, Geneva, 2021).
-
- United Nations Children’s Fund (UNICEF). One is too many: Ending child deaths from pneumonia and diarrhea. (UNICEF, New York, 2016).
-
- World Health Organization. Daily iron supplementation in children 6–23 months of age. e-Library of Evidence for Nutrition Actions (eLENA), Vol. 2016 (World Health Organization, Geneva, 2016).
-
- WHO Guidelines Approved by the Guidelines Review Committee. Use of multiple micronutrient powders for point-of-use fortification of foods consumed by infants and young children aged 6–23 months and children aged 2–12 years (World Health Organization, Geneva, 2016). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

