Quantitative proteomics and phosphoproteomics profiling of meiotic divisions in the fission yeast Schizosaccharomyces pombe
- PMID: 39367033
- PMCID: PMC11452395
- DOI: 10.1038/s41598-024-74523-0
Quantitative proteomics and phosphoproteomics profiling of meiotic divisions in the fission yeast Schizosaccharomyces pombe
Abstract
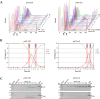
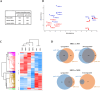
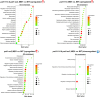
In eukaryotes, chromosomal DNA is equally distributed to daughter cells during mitosis, whereas the number of chromosomes is halved during meiosis. Despite considerable progress in understanding the molecular mechanisms that regulate mitosis, there is currently a lack of complete understanding of the molecular mechanisms regulating meiosis. Here, we took advantage of the fission yeast Schizosaccharomyces pombe, for which highly synchronous meiosis can be induced, and performed quantitative proteomics and phosphoproteomics analyses to track changes in protein expression and phosphorylation during meiotic divisions. We compared the proteomes and phosphoproteomes of exponentially growing mitotic cells with cells harvested around meiosis I, or meiosis II in strains bearing either the temperature-sensitive pat1-114 allele or conditional ATP analog-sensitive pat1-as2 allele of the Pat1 kinase. Comparing pat1-114 with pat1-as2 also allowed us to investigate the impact of elevated temperature (25 °C versus 34 °C) on meiosis, an issue that sexually reproducing organisms face due to climate change. Using TMTpro 18plex labeling and phosphopeptide enrichment strategies, we performed quantification of a total of 4673 proteins and 7172 phosphosites in S. pombe. We found that the protein level of 2680 proteins and the rate of phosphorylation of 4005 phosphosites significantly changed during progression of S. pombe cells through meiosis. The proteins exhibiting changes in expression and phosphorylation during meiotic divisions were represented mainly by those involved in the meiotic cell cycle, meiotic recombination, meiotic nuclear division, meiosis I, centromere clustering, microtubule cytoskeleton organization, ascospore formation, organonitrogen compound biosynthetic process, carboxylic acid metabolic process, gene expression, and ncRNA processing, among others. In summary, our findings provide global overview of changes in the levels and phosphorylation of proteins during progression of S. pombe cells through meiosis at normal and elevated temperatures, laying the groundwork for further elucidation of the functions and importance of specific proteins and their phosphorylation in regulating meiotic divisions in this yeast.
Keywords: Schizosaccharomyces pombe; Elevated temperature; Meiosis; Pat1 protein kinase; Phosphoproteome; Proteome; Quantitative mass spectrometry.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
ATP analog-sensitive Pat1 protein kinase for synchronous fission yeast meiosis at physiological temperature.Cell Cycle. 2012 Apr 15;11(8):1626-33. doi: 10.4161/cc.20052. Epub 2012 Apr 15. Cell Cycle. 2012. PMID: 22487684 Free PMC article.
-
SILAC-Based Proteomic Analysis of Meiosis in the Fission Yeast Schizosaccharomyces pombe.Methods Mol Biol. 2023;2603:19-29. doi: 10.1007/978-1-0716-2863-8_2. Methods Mol Biol. 2023. PMID: 36370267
-
Meiosis induced by inactivation of Pat1 kinase proceeds with aberrant nuclear positioning of centromeres in the fission yeast Schizosaccharomyces pombe.Genes Cells. 2004 Aug;9(8):671-84. doi: 10.1111/j.1356-9597.2004.00760.x. Genes Cells. 2004. PMID: 15298676
-
CDK Regulation of Meiosis: Lessons from S. cerevisiae and S. pombe.Genes (Basel). 2020 Jun 29;11(7):723. doi: 10.3390/genes11070723. Genes (Basel). 2020. PMID: 32610611 Free PMC article. Review.
-
Homologous chromosome pairing in Schizosaccharomyces pombe.Yeast. 2006 Oct 15;23(13):977-89. doi: 10.1002/yea.1403. Yeast. 2006. PMID: 17072890 Review.
Cited by
-
Molecular and Biophysical Perspectives on Dormancy Breaking: Lessons from Yeast Spore.Biomolecules. 2025 May 11;15(5):701. doi: 10.3390/biom15050701. Biomolecules. 2025. PMID: 40427594 Free PMC article. Review.
-
Roles for the canonical polarity machinery in the de novo establishment of polarity in budding yeast spores.bioRxiv [Preprint]. 2025 Jan 6:2024.08.29.610423. doi: 10.1101/2024.08.29.610423. bioRxiv. 2025. Update in: Mol Biol Cell. 2025 Mar 01;36(3):ar28. doi: 10.1091/mbc.E24-07-0303. PMID: 39257763 Free PMC article. Updated. Preprint.
-
Roles for the canonical polarity machinery in the de novo establishment of polarity in budding yeast spores.Mol Biol Cell. 2025 Mar 1;36(3):ar28. doi: 10.1091/mbc.E24-07-0303. Epub 2025 Jan 22. Mol Biol Cell. 2025. PMID: 39841544 Free PMC article.
References
-
- Okaz, E. et al. Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/CAma1. Cell. 151 (3), 603–618 (2012). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

