ARF1 compartments direct cargo flow via maturation into recycling endosomes
- PMID: 39367144
- PMCID: PMC11567898
- DOI: 10.1038/s41556-024-01518-4
ARF1 compartments direct cargo flow via maturation into recycling endosomes
Abstract
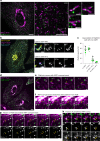
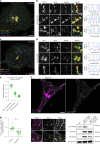
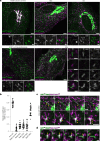
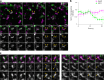
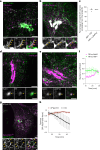
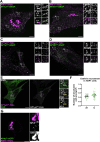
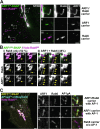
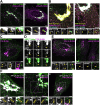
Cellular membrane homoeostasis is maintained via a tightly regulated membrane and cargo flow between organelles of the endocytic and secretory pathways. Adaptor protein complexes (APs), which are recruited to membranes by the small GTPase ARF1, facilitate cargo selection and incorporation into trafficking intermediates. According to the classical model, small vesicles would facilitate bi-directional long-range transport between the Golgi, endosomes and plasma membrane. Here we revisit the intracellular organization of the vesicular transport machinery using a combination of CRISPR-Cas9 gene editing, live-cell high temporal (fast confocal) or spatial (stimulated emission depletion) microscopy as well as correlative light and electron microscopy. We characterize tubulo-vesicular ARF1 compartments that harbour clathrin and different APs. Our findings reveal two functionally different classes of ARF1 compartments, each decorated by a different combination of APs. Perinuclear ARF1 compartments facilitate Golgi export of secretory cargo, while peripheral ARF1 compartments are involved in endocytic recycling downstream of early endosomes. Contrary to the classical model of long-range vesicle shuttling, we observe that ARF1 compartments shed ARF1 and mature into recycling endosomes. This maturation process is impaired in the absence of AP-1 and results in trafficking defects. Collectively, these data highlight a crucial role for ARF1 compartments in post-Golgi sorting.
© 2024. The Author(s).
Conflict of interest statement
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
- SFB958 Project A25/Deutsche Forschungsgemeinschaft (German Research Foundation)
- TRR186 Project A20/Deutsche Forschungsgemeinschaft (German Research Foundation)
- TRR186 project A08/Deutsche Forschungsgemeinschaft (German Research Foundation)
- HA 2686/24-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- TRR186 project Z02/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

