Multiomic profiling identifies predictors of survival in African American patients with acute myeloid leukemia
- PMID: 39367245
- PMCID: PMC11549055
- DOI: 10.1038/s41588-024-01929-x
Multiomic profiling identifies predictors of survival in African American patients with acute myeloid leukemia
Abstract
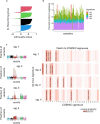
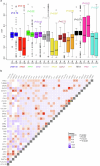
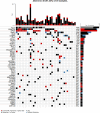
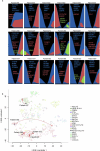
Genomic profiles and prognostic biomarkers in patients with acute myeloid leukemia (AML) from ancestry-diverse populations are underexplored. We analyzed the exomes and transcriptomes of 100 patients with AML with genomically confirmed African ancestry (Black; Alliance) and compared their somatic mutation frequencies with those of 323 self-reported white patients with AML, 55% of whom had genomically confirmed European ancestry (white; BeatAML). Here we find that 73% of 162 gene mutations recurrent in Black patients, including a hitherto unreported PHIP alteration detected in 7% of patients, were found in one white patient or not detected. Black patients with myelodysplasia-related AML were younger than white patients suggesting intrinsic and/or extrinsic dysplasia-causing stressors. On multivariable analyses of Black patients, NPM1 and NRAS mutations were associated with inferior disease-free and IDH1 and IDH2 mutations with reduced overall survival. Inflammatory profiles, cell type distributions and transcriptional profiles differed between Black and white patients with NPM1 mutations. Incorporation of ancestry-specific risk markers into the 2022 European LeukemiaNet genetic risk stratification changed risk group assignment for one-third of Black patients and improved their outcome prediction.
© 2024. The Author(s).
Conflict of interest statement
C.J.W. is a consultant for Vigeo Therapeutics and is employed by Karyopharm Therapeutics; he has ownership interest in Karyopharm Therapeutics and Bristol Myers Squibb Co. A.S.M. has served in a consulting or advisory role for AbbVie, Bristol Meyers Squibb, Novartis and Treadwell Therapeutics; he has served on a data monitoring safety committee for Daiichi Sankyo and Foghorn Therapeutics and currently serves as a Senior Medical Director for the Leukemia and Lymphoma Society Beat AML Study. J.S.B. is a consultant and advisory board member for AbbVie, AstraZeneca, Syndax, INNATE and KITE. B.L.P. has received honoraria from Jazz Pharmaceuticals, Novartis and Pfizer. J.F.K. has received honoraria from Gilead, Magellan and Novartis; consulting fees from Gilead, Magellan, Novartis, Pharmacyclics and Seattle Genetics; institutional research funding from Boehringer Ingelheim, Cantex, Erytech and Millennium; and travel support from Gilead, Novartis and Seattle Genetics. R.M.S. has served on independent data safety monitoring committees for trials supported by Celgene, Takeda and Argenx; has consulted for AbbVie, Actinium, Agios, Amgen, Arog, Astellas, AstraZeneca, BioLineRx, Celgene, Daiichi Sankyo, Fujifilm, Janssen, Juno, Macrogenics, Novartis, Ono, Orsenix, Pfizer, Roche, Stemline Therapeutics, Sumitomo, Takeda and Trovagene; and has received research support (to his institution) for clinical trials sponsored by AbbVie, Agios, Arog and Novartis. E.D.P. has received grants from the Merck Foundation and Pfizer. M.L.G. holds research contracts and consults with Bridge Medicines and holds stock options for SeqRx. M.F.B. has a consultancy role with AstraZeneca, Eli Lilly and Paige.AI and has received research support from Boundless Bio. J.C.B. has a consultancy and advisory role with Syndax, Novartis and Vincera; research funding from Pharmacyclics, an AbbVie company, Genentech, Janssen and Acerta; and ownership for Vincera. E.R.M. has received research support (to her institution) from Pfizer, the Merck Foundation, Genentech, Guardant Health and Astra Zeneca; and has served as an advisory board member for GSK and Merck, and as member of the Board of Directors of the Alliance Foundation and American Association for Cancer Research. R.L.L. is on the supervisory board of QIAGEN and is a scientific adviser to Imago, Mission Bio, Syndax, Zentalis Pharmaceuticals, Ajax, Bakx, Auron, Prelude, C4 Therapeutics and Isoplexis, for which he receives equity support. R.L.L. receives research support from Ajax and Abbvie and has consulted for Incyte, Janssen, MorphoSys and Novartis. He has received honoraria from AstraZeneca and Incyte for invited lectures. A.-K.E. has received a research grant from Novartis, and an honorarium from AstraZeneca for serving on their Diversity, Equity and Inclusion Advisory Board; her spouse has ownership interest in Karyopharm Therapeutics. The other authors declare no completing interests.
Figures

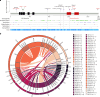


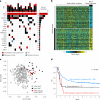
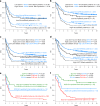

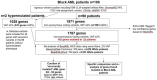


References
-
- Döhner, H., Weisdorf, D. J. & Bloomfield, C. D. Acute myeloid leukemia. N. Engl. J. Med.373, 1136–1152 (2015). - PubMed
MeSH terms
Substances
Grants and funding
- 2023YIA-5335030875/Conquer Cancer Foundation (Conquer Cancer Foundation of the American Society of Clinical Oncology)
- T32 CA117846/CA/NCI NIH HHS/United States
- UG1 CA233324/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R35 CA197594/CA/NCI NIH HHS/United States
- R01 CA283574-01/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- UG1 CA189850/CA/NCI NIH HHS/United States
- UG1 CA233180/CA/NCI NIH HHS/United States
- R50 CA275927/CA/NCI NIH HHS/United States
- UG1 CA189824/CA/NCI NIH HHS/United States
- U10 CA180867/CA/NCI NIH HHS/United States
- R35 CA197734/CA/NCI NIH HHS/United States
- R01 CA262496/CA/NCI NIH HHS/United States
- UG1 CA233290/CA/NCI NIH HHS/United States
- P50 CA140158/CA/NCI NIH HHS/United States
- R01 CA284595/CA/NCI NIH HHS/United States
- UG1 CA233331/CA/NCI NIH HHS/United States
- U10 CA180821/CA/NCI NIH HHS/United States
- R01 CA283574/CA/NCI NIH HHS/United States
- U10 CA180882/CA/NCI NIH HHS/United States
- R01 LM013879/LM/NLM NIH HHS/United States
- UG1 CA233338/CA/NCI NIH HHS/United States
- U24 CA196171/CA/NCI NIH HHS/United States
- P30 CA016058/CA/NCI NIH HHS/United States
- UG1 CA233327/CA/NCI NIH HHS/United States
- R01 CA284595-01/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- U10 CA180888/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

