Noninvasive, microbiome-based diagnosis of inflammatory bowel disease
- PMID: 39367251
- PMCID: PMC11645270
- DOI: 10.1038/s41591-024-03280-4
Noninvasive, microbiome-based diagnosis of inflammatory bowel disease
Abstract
Despite recent progress in our understanding of the association between the gut microbiome and inflammatory bowel disease (IBD), the role of microbiome biomarkers in IBD diagnosis remains underexplored. Here we developed a microbiome-based diagnostic test for IBD. By utilization of metagenomic data from 5,979 fecal samples with and without IBD from different geographies and ethnicities, we identified microbiota alterations in IBD and selected ten and nine bacterial species for construction of diagnostic models for ulcerative colitis and Crohn's disease, respectively. These diagnostic models achieved areas under the curve >0.90 for distinguishing IBD from controls in the discovery cohort, and maintained satisfactory performance in transethnic validation cohorts from eight populations. We further developed a multiplex droplet digital polymerase chain reaction test targeting selected IBD-associated bacterial species, and models based on this test showed numerically higher performance than fecal calprotectin in discriminating ulcerative colitis and Crohn's disease from controls. Here we discovered universal IBD-associated bacteria and show the potential applicability of a multibacteria biomarker panel as a noninvasive tool for IBD diagnosis.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: S.C.N. has served as an advisory board member for Pfizer, Ferring, Janssen and Abbvie and received honoraria as a speaker for Ferring, Tillotts, Menarini, Janssen, Abbvie and Takeda; has received research grants through her affiliated institutions from Olympus, Ferring and Abbvie; is a founder member, nonexecutive director, nonexecutive scientific advisor and shareholder of GenieBiome Ltd; and receives patent royalties through her affiliated institutions. F.K.L.C. is a Board Member of CUHK Medical Centre; is a cofounder, nonexecutive Board Chairman, nonexecutive scientific advisor, Honorary Chief Medical Officer and shareholder of GenieBiome Ltd; receives patent royalties through his affiliated institutions; and has received fees as an advisor and honoraria as a speaker for Eisai Co. Ltd, AstraZeneca, Pfizer Inc., Takeda Pharmaceutical Co. and Takeda (China) Holdings Co. Ltd. Q. Su and Z.X. are Scientists (Diagnostics) of GenieBiome Ltd. W.T. is Consultant (Regulatory Affairs) of GenieBiome Ltd. J. Zhang is Chief Scientist (Diagnostics) of GenieBiome Ltd. J. Zheng, W.T., J. Zhang, F.K.L.C. and S.C.N. are named inventors of patent applications held by MagIC that cover the therapeutic and diagnostic use of microbiome related to IBD (nos. 63/562,232; 63/675,266; 63/689,864 USA, 2024). C.N.B. is supported by the Bingham Chair in Gastroenterology; has served on advisory Boards for AbbVie Canada, Amgen Canada, Bristol Myers Squibb Canada, Ferring Canada, JAMP Pharmaceuticals, Lilly Canada, Janssen Canada, Pendopharm Canada, Sandoz Canada, Takeda Canada and Pfizer Canada; has received educational grants from Abbvie Canada, Bristol Myers Squibb Canada, Ferring Canada, organon Canada, Pfizer Canada, Takeda Canada, Boston Scientific and Janssen Canada; has served on a Speaker’s panel for Abbvie Canada, Janssen Canada, Pfizer Canada and Takeda Canada; and has received research funding from Abbvie Canada, Amgen Canada, Sandoz Canada, Takeda Canada and Pfizer Canada. D.T.R. has received grant support from Takeda and has served as a consultant for Abbvie, Altrubio, Amgen, Bausch Health, Bristol Myers Squibb, Connect BioPharma, Ferring Pharma, Image Analysis Group, Iterative Health, Janssen Pharmaceuticals, Lilly, Merck, Pfizer, Prometheus Biosciences (now Merck), Reistone Biopharma, USA, Takeda and Trellus Health. E.B.C. is Founder and Chief Scientific Officer of Gateway Biome, Inc. M.M. has received research grants from Soho Flordis International Australia Research, Bayer Steigerwald Arzneimittelwerk (Bayer Consumer Health) and Yakult-Nature Global Grant for Gut Health; Speaker’s honoraria and travel sponsorship from Janssen Australia; consultancy fees from Bayer Steigerwald Arzneimittelwerk (Bayer Consumer Health), Sanofi Australia and Danone-Nutricia Australia; Speaker honoraria and travel sponsorship from Perfect Company (China); and travel sponsorship from Yakult Inc. (Japan); is coinventor of PCT/AU2022/050556 ‘Diagnostic marker for functional gastrointestinal disorders’ from the University of Newcastle and UniQuest (University of Queensland), and US20110076356 A1 ‘Novel Fibro-biotic bacterium isolate’ via the US Department of Agriculture; acknowledges funding from NHMRC Australia, Australian Research Council, Princess Alexandra Hospital Research Foundation, Medical Research Futures Fund of Australia, Helmsley Charitable Trust via the Australasian Gastrointestinal Research Foundation and the United States Department of Defense; and serves on the science advisory board (nonremunerated) for GenieBiome, Hong Kong. The other authors declare no competing interests.
Figures








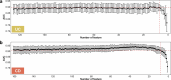

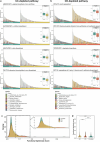


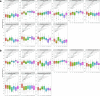


References
-
- Ng, S. C. et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet390, 2769–2778 (2017). - PubMed
-
- Noble, A. J., Nowak, J. K., Adams, A. T., Uhlig, H. H. & Satsangi, J. Defining interactions between the genome, epigenome, and the environment in inflammatory bowel disease: progress and prospects. Gastroenterology165, 44–60 (2023). - PubMed
-
- Ananthakrishnan, A. N. et al. Environmental triggers in IBD: a review of progress and evidence. Nat. Rev. Gastroenterol. Hepatol.15, 39–49 (2018). - PubMed
MeSH terms
Substances
Grants and funding
- R4030-22/Research Grants Council, University Grants Committee (RGC, UGC)
- 14121322/Research Grants Council, University Grants Committee (RGC, UGC)
- 2017PG-IBD003/Leona M. and Harry B. Helmsley Charitable Trust (Helmsley Charitable Trust)
- 10210816/Food and Health Bureau of the Government of the Hong Kong Special Administrative Region | Health and Medical Research Fund (HMRF)
- 82100573/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical

