A decade of dinoflagellate genomics illuminating an enigmatic eukaryote cell
- PMID: 39367346
- PMCID: PMC11453091
- DOI: 10.1186/s12864-024-10847-5
A decade of dinoflagellate genomics illuminating an enigmatic eukaryote cell
Abstract
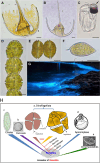
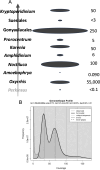
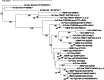
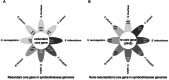
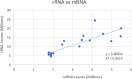
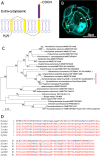
Dinoflagellates are a remarkable group of protists, not only for their association with harmful algal blooms and coral reefs but also for their numerous characteristics deviating from the rules of eukaryotic biology. Genome research on dinoflagellates has lagged due to their immense genome sizes in most species (~ 1-250 Gbp). Nevertheless, the last decade marked a fruitful era of dinoflagellate genomics, with 27 genomes sequenced and many insights attained. This review aims to synthesize information from these genomes, along with other omic data, to reflect on where we are now in understanding dinoflagellates and where we are heading in the future. The most notable insights from the decade-long genomics work include: (1) dinoflagellate genomes have been expanded in multiple times independently, probably by a combination of rampant retroposition, accumulation of repetitive DNA, and genome duplication; (2) Symbiodiniacean genomes are highly divergent, but share about 3,445 core unigenes concentrated in 219 KEGG pathways; (3) Most dinoflagellate genes are encoded unidirectionally and are not intron-poor; (4) The dinoflagellate nucleus has undergone extreme evolutionary changes, including complete or nearly complete loss of nucleosome and histone H1, and acquisition of dinoflagellate viral nuclear protein (DVNP); (5) Major basic nuclear protein (MBNP), histone-like protein (HLP), and bacterial HU-like protein (HCc) belong to the same protein family, and MBNP can be the unifying name; (6) Dinoflagellate gene expression is regulated by poorly understood mechanisms, but microRNA and other epigenetic mechanisms are likely important; (7) Over 50% of dinoflagellate genes are "dark" and their functions remain to be deciphered using functional genetics; (8) Initial insights into the genomic basis of parasitism and mutualism have emerged. The review then highlights functionally unique and interesting genes. Future research needs to obtain a finished genome, tackle large genomes, characterize the unknown genes, and develop a quantitative molecular ecological model for addressing ecological questions.
Keywords: Dinoflagellate; Gene regulation; Genome expansion; Histone; MicroRNA (miRNA); Nuclear protein; Omics; Parasitic; Retroposition; Symbiotic.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Fensome RA. A classification of living and fossil dinoflagellates. Micropaleontology Spec Publ. 1993;7:351.
-
- Gómez F. A checklist and classification of living dinoflagellates (Dinoflagellata, Alveolata). Cicimar Oceánides. 2012;27(1):65–140. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

