Discovery of a selective cytochrome P450 4A inhibitor for the treatment of metabolic dysfunction-associated fatty liver disease
- PMID: 39367660
- PMCID: PMC11452733
- DOI: 10.1002/ctm2.1816
Discovery of a selective cytochrome P450 4A inhibitor for the treatment of metabolic dysfunction-associated fatty liver disease
Conflict of interest statement
The authors declare no conflict of interest.
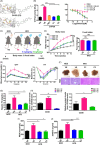
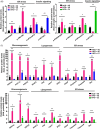
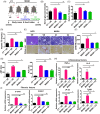
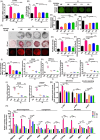
Figures

References
-
- Eslam M, Newsome PN, Sarin SK, et al. A new definition for metabolic dysfunction‐associated fatty liver disease: an international expert consensus statement. Journal of Hepatology. 2020;73(1):202‐209. - PubMed
-
- Eslam M, Sanyal AJ, George J, et al. MAFLD: a consensus‐driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158(7):1999‐2014. e1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
