DCAF13 promotes ovarian cancer progression by activating FRAS1-mediated FAK signaling pathway
- PMID: 39367995
- PMCID: PMC11455852
- DOI: 10.1007/s00018-024-05446-2
DCAF13 promotes ovarian cancer progression by activating FRAS1-mediated FAK signaling pathway
Abstract
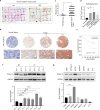
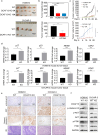
Cullin-RING ubiquitin ligase 4 (CRL4) is closely correlated with the incidence and progression of ovarian cancer. DDB1- and CUL4-associated factor 13 (DCAF13), a substrate-recognition protein in the CRL4 E3 ubiquitin ligase complex, is involved in the occurrence and development of ovarian cancer. However, its precise function and the underlying molecular mechanism in this disease remain unclear. In this study, we confirmed that DCAF13 is highly expressed in human ovarian cancer and its expression is negatively correlated with the overall survival rate of patients with ovarian cancer. We then used CRISPR/Cas9 to knockout DCAF13 and found that its deletion significantly inhibited the proliferation, colony formation, and migration of human ovarian cancer cells. In addition, DCAF13 deficiency inhibited tumor proliferation in nude mice. Mechanistically, CRL4-DCAF13 targeted Fraser extracellular matrix complex subunit 1 (FRAS1) for polyubiquitination and proteasomal degradation. FRAS1 influenced the proliferation and migration of ovarian cancer cell through induction of the focal adhesion kinase (FAK) signaling pathway. These findings collectively show that DCAF13 is an important oncogene that promotes tumorigenesis in ovarian cancer cells by mediating FRAS1/FAK signaling. Our findings provide a foundation for the development of targeted therapeutics for ovarian cancer.
Keywords: CRL4 E3 ubiquitin ligase; DCAF13; FAK; FRAS1; Ovarian cancer.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249. 10.3322/caac.21660 - PubMed
-
- Menon U, Gentry-Maharaj A, Burnell M, Singh N, Ryan A, Karpinskyj C et al (2021) Ovarian cancer population screening and mortality after long-term follow-up in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet 397(10290):2182–2193. 10.1016/S0140-6736(21)00731-5 - PMC - PubMed
-
- Van Zyl B, Tang D, Bowden NA (2018) Biomarkers of platinum resistance in ovarian cancer: what can we use to improve treatment. Endocr Relat Cancer 25(5):R303–R318. 10.1530/ERC-17-0336 - PubMed
MeSH terms
Substances
Grants and funding
- 31871402/National Natural Science Foundation of China
- LY21H160047/Natural Science Foundation of Zhejiang Province
- LGD21H160003/Natural Science Foundation of Zhejiang Province
- LQ23C070001/Natural Science Foundation of Zhejiang Province
- LY17H160060/Natural Science Foundation of Zhejiang Province
- Z20H160031/Natural Science Foundation of Zhejiang Province
- LGF20H160031/Natural Science Foundation of Zhejiang Province
- LGD22H030004/Natural Science Foundation of Zhejiang Province
- 12.2018/Zhejiang Provincial Foreign Expert Grant
- 12.2019/Jiaxing Key Laboratory for Photonanomedicine and Experimental Therapeutics
- KWF, project 10666/Dutch Cancer Foundation
- 6.2021/Jiaxing talent pioneer innovation team
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

