CDK12 loss drives prostate cancer progression, transcription-replication conflicts, and synthetic lethality with paralog CDK13
- PMID: 39368479
- PMCID: PMC11513839
- DOI: 10.1016/j.xcrm.2024.101758
CDK12 loss drives prostate cancer progression, transcription-replication conflicts, and synthetic lethality with paralog CDK13
Abstract
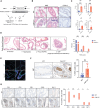
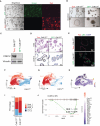
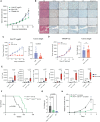
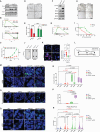
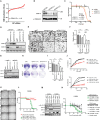
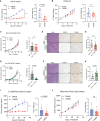
Biallelic loss of cyclin-dependent kinase 12 (CDK12) defines a metastatic castration-resistant prostate cancer (mCRPC) subtype. It remains unclear, however, whether CDK12 loss drives prostate cancer (PCa) development or uncovers pharmacologic vulnerabilities. Here, we show Cdk12 ablation in murine prostate epithelium is sufficient to induce preneoplastic lesions with lymphocytic infiltration. In allograft-based CRISPR screening, Cdk12 loss associates positively with Trp53 inactivation but negatively with Pten inactivation. Moreover, concurrent Cdk12/Trp53 ablation promotes proliferation of prostate-derived organoids, while Cdk12 knockout in Pten-null mice abrogates prostate tumor growth. In syngeneic systems, Cdk12/Trp53-null allografts exhibit luminal morphology and immune checkpoint blockade sensitivity. Mechanistically, Cdk12 inactivation mediates genomic instability by inducing transcription-replication conflicts. Strikingly, CDK12-mutant organoids and patient-derived xenografts are sensitive to inhibition or degradation of the paralog kinase, CDK13. We therein establish CDK12 as a bona fide tumor suppressor, mechanistically define how CDK12 inactivation causes genomic instability, and advance a therapeutic strategy for CDK12-mutant mCRPC.
Keywords: CDK12; CDK13; Cdk12 knockout; R-loops; paralog-based synthetic lethality; prostate cancer; transcription-replication conflicts.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.M.C. co-founded and serves on scientific advisory boards (SABs) of Lynx Dx, Flamingo Therapeutics, Medsyn Pharma, Oncopia Therapeutics, and Esanik Therapeutics. A.M.C. is an advisor to Aurigene Oncology Limited, Proteovant, Tempus, Rappta, and Ascentage. C.J.L. received research funding from AstraZeneca, Merck KGaA, Artios, and NeoPhore and consultancy, SAB membership, or honoraria payments from FoRx, Syncona, Sun Pharma, Gerson Lehrman Group, Merck KGaA, Vertex, AstraZeneca, Tango, 3rd Rock, Ono Pharma, Artios, Abingworth, Tesselate, Dark Blue Therapeutics, Pontifax, Astex, NeoPhore, Glaxo Smith Kline, and Dawn Bioventures. C.J.L. has stock in Tango, Ovibio, Hysplex, and Tesselate. C.J.L. is named inventor on patents describing use of DNA repair inhibitors and stands to gain from their development and use. J.C. is an advisor for Exai Bio. F.Y.F. has served on SAB or received consulting fees from Astellas, Bayer, Celgene, Clovis Oncology, Janssen, Genentech Roche, Myovant, Roivant, Sanofi, and Blue Earth Diagnostics. F.Y.F. is also an SAB member for Artera, ClearNote Genomics, Serimmune, and BMS (Microenvironment Division). K.D. is an advisor for Kinoteck Therapeutics and has received financial support from Livzon Pharmaceutical Group. Patents for CDK12/13 degraders/inhibitors used here have been filed by the University of Michigan and Shanghai Institute of Organic Chemistry, with A.M.C., K.D., X.W., J.Y., Y. Chang, and J.C.T. as co-inventors.
Figures


Update of
-
CDK12 Loss Promotes Prostate Cancer Development While Exposing Vulnerabilities to Paralog-Based Synthetic Lethality.bioRxiv [Preprint]. 2024 Mar 21:2024.03.20.585990. doi: 10.1101/2024.03.20.585990. bioRxiv. 2024. Update in: Cell Rep Med. 2024 Oct 15;5(10):101758. doi: 10.1016/j.xcrm.2024.101758. PMID: 38562774 Free PMC article. Updated. Preprint.
References
-
- Chou J., Quigley D.A., Robinson T.M., Feng F.Y., Ashworth A. Transcription-Associated Cyclin-Dependent Kinases as Targets and Biomarkers for Cancer Therapy. Cancer Discov. 2020;10:351–370. doi: 10.1158/2159-8290.CD-19-0528. - DOI - PubMed
-
- Zhang T., Kwiatkowski N., Olson C.M., Dixon-Clarke S.E., Abraham B.J., Greifenberg A.K., Ficarro S.B., Elkins J.M., Liang Y., Hannett N.M., et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol. 2016;12:876–884. doi: 10.1038/nchembio.2166. - DOI - PMC - PubMed
-
- Blazek D., Kohoutek J., Bartholomeeusen K., Johansen E., Hulinkova P., Luo Z., Cimermancic P., Ule J., Peterlin B.M. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25:2158–2172. doi: 10.1101/gad.16962311. - DOI - PMC - PubMed
-
- Cheng S.W.G., Kuzyk M.A., Moradian A., Ichu T.A., Chang V.C.D., Tien J.F., Vollett S.E., Griffith M., Marra M.A., Morin G.B. Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. Mol. Cell Biol. 2012;32:4691–4704. doi: 10.1128/MCB.06267-11. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

