Pan-cancer γδ TCR analysis uncovers clonotype diversity and prognostic potential
- PMID: 39368482
- PMCID: PMC11513832
- DOI: 10.1016/j.xcrm.2024.101764
Pan-cancer γδ TCR analysis uncovers clonotype diversity and prognostic potential
Abstract
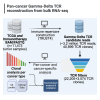
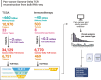
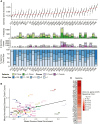
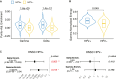
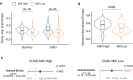
Gamma-delta T cells (γδ T cells) play a crucial role in both innate and adaptive immunity within tumors, yet their presence and prognostic value in cancer remain underexplored. This study presents a large-scale analysis of γδ T cell receptor (γδ TCR) reads from 11,000 tumor samples spanning 33 cancer types, utilizing the TRUST4 algorithm. Our findings reveal extensive diversity in γδ TCR clonality and gene expression, underscoring the potential of γδ T cells as prognostic biomarkers in various cancers. We further demonstrate the utility of TCR gamma (TRG) and delta (TRD) gene expression from standard RNA-sequencing (RNA-seq) data. This comprehensive dataset offers a valuable resource for advancing γδ T cell research, with implications for enhanced immunotherapy approaches or alternative therapeutic strategies. Additionally, our centralized database supports translational research into the therapeutic significance of γδ T cells.
Keywords: T cell receptor; TCR clonality; TRUST4; cancer immunotherapy; gamma-delta T cells; immune repertoire; prognostic biomarkers.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.R.C.-G. is a consultant for Anixa Biosciences and Alloy Therapeutics; holds stock options in Compass Therapeutics, Anixa Biosciences, and Alloy Therapeutics; has a patent licensed by Anixa Biosciences; owns intellectual property filed with Compass Therapeutics; and is a co-founder of Cellepus Therapeutics, a CAR T cell company.
Figures


References
-
- Corvaisier M., Moreau-Aubry A., Diez E., Bennouna J., Mosnier J.-F., Scotet E., Bonneville M., Jotereau F. Vγ9Vδ2 T cell response to colon carcinoma cells. J. Immunol. 2005;175:5481–5488. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

