Clinical and immune responses to neoadjuvant fulvestrant with or without enzalutamide in ER+/Her2- breast cancer
- PMID: 39368973
- PMCID: PMC11455938
- DOI: 10.1038/s41523-024-00697-5
Clinical and immune responses to neoadjuvant fulvestrant with or without enzalutamide in ER+/Her2- breast cancer
Abstract
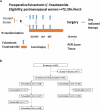
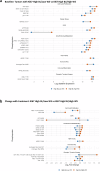
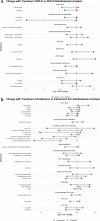
Most ER+ breast cancers (BC) express androgen receptors (AR). This randomized phase II trial of 4 months of neoadjuvant fulvestrant (Fulv) alone or with enzalutamide (Combo) assessed whether adding AR blockade to Fulv would limit residual tumor at the time of surgery, as measured by modified preoperative endocrine predictive index (PEPI) score. Eligible patients were women with ER+/HER2- primary BC cT2 or greater. Stratification factors were clinical node and T-stage. Fresh tumor biopsies were required at study entry, after 4 weeks on therapy (W5), and at surgery. Laboratory analyses on tumors included immunochemistry (IHC) for ER/PR/AR/GR and Ki67 protein, evaluation of gene expression, multiplex for myeloid lineage immune cells, reverse-phase protein array, and plasma metabolomic analyses. Of 69 consented patients, 59 were evaluable. Toxicity was as expected with endocrine therapy. Combo achieved PEPI = 0 more frequently (24%: 8/33) than Fulv (8%: 2/26). Ki67 was ≤10% across arms by W5 in 76% of tumors. Activation of mTOR pathway proteins was elevated in tumors with poor Ki67 response. Tumors in both arms showed decreased estrogen-regulated and cell division gene sets, while Combo arm tumors uniquely exhibited enrichment of immune activation gene sets, including interferon gamma, complement, inflammation, antigen processing, and B and T cell activation. Multiplex IHC showed significantly reduced tumor-associated macrophages and CD14+/HLADR-/CD68- MDSCs in Combo tumors at W5. In summary, Combo tumors showed a higher PEPI = 0 response, Ki67 response, and more activated tumor immune microenvironment than Fulv. The odds of response were 4.6-fold higher for patients with ILC versus IDC. (Trial registration: This trial is registered at Clinicaltrials.gov ( https://www.clinicaltrials.gov/study/NCT02955394?id=16-1042&rank=1 ). The trial registration number is NCT02955394. The full trial protocol is available under Study Details at the Clinicaltrials.gov link provided).
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Ricciardelli, C. et al. The magnitude of androgen receptor positivity in breast cancer is critical for reliable prediction of disease outcome. Clin. Cancer Res.24, 2328–2341 (2018). - PubMed
-
- Cao, L. et al. A high AR:ERα or PDEF:ERα ratio predicts a sub-optimal response to tamoxifen therapy in ERα-positive breast cancer. Cancer Chemother. Pharmacol.84, 609–620 (2019). - PubMed
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

