Metabolic regulation of mitochondrial morphologies in pancreatic beta cells: coupling of bioenergetics and mitochondrial dynamics
- PMID: 39369076
- PMCID: PMC11455970
- DOI: 10.1038/s42003-024-06955-3
Metabolic regulation of mitochondrial morphologies in pancreatic beta cells: coupling of bioenergetics and mitochondrial dynamics
Abstract
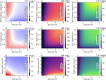
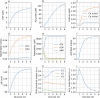
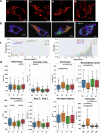
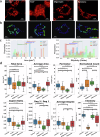
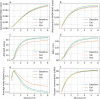
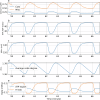
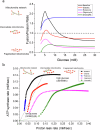
Cellular bioenergetics and mitochondrial dynamics are crucial for the secretion of insulin by pancreatic beta cells in response to elevated levels of blood glucose. To elucidate the interactions between energy production and mitochondrial fission/fusion dynamics, we combine live-cell mitochondria imaging with biophysical-based modeling and graph-based network analysis. The aim is to determine the mechanism that regulates mitochondrial morphology and balances metabolic demands in pancreatic beta cells. A minimalistic differential equation-based model for beta cells is constructed that includes glycolysis, oxidative phosphorylation, calcium dynamics, and fission/fusion dynamics, with ATP synthase flux and proton leak flux as main regulators of mitochondrial dynamics. The model shows that mitochondrial fission occurs in response to hyperglycemia, starvation, ATP synthase inhibition, uncoupling, and diabetic conditions, in which the rate of proton leakage exceeds the rate of mitochondrial ATP synthesis. Under these metabolic challenges, the propensities of tip-to-tip fusion events simulated from the microscopy images of the mitochondrial networks are lower than those in the control group and prevent the formation of mitochondrial networks. The study provides a quantitative framework that couples bioenergetic regulation with mitochondrial dynamics, offering insights into how mitochondria adapt to metabolic challenges.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

