Uncovering the small proteome of Methanosarcina mazei using Ribo-seq and peptidomics under different nitrogen conditions
- PMID: 39370430
- PMCID: PMC11456600
- DOI: 10.1038/s41467-024-53008-8
Uncovering the small proteome of Methanosarcina mazei using Ribo-seq and peptidomics under different nitrogen conditions
Abstract
The mesophilic methanogenic archaeal model organism Methanosarcina mazei strain Gö1 is crucial for climate and environmental research due to its ability to produce methane. Here, we establish a Ribo-seq protocol for M. mazei strain Gö1 under two growth conditions (nitrogen sufficiency and limitation). The translation of 93 previously annotated and 314 unannotated small ORFs, coding for proteins ≤ 70 amino acids, is predicted with high confidence based on Ribo-seq data. LC-MS analysis validates the translation for 62 annotated small ORFs and 26 unannotated small ORFs. Epitope tagging followed by immunoblotting analysis confirms the translation of 13 out of 16 selected unannotated small ORFs. A comprehensive differential transcription and translation analysis reveals that 29 of 314 unannotated small ORFs are differentially regulated in response to nitrogen availability at the transcriptional and 49 at the translational level. A high number of reported small RNAs are emerging as dual-function RNAs, including sRNA154, the central regulatory small RNA of nitrogen metabolism. Several unannotated small ORFs are conserved in Methanosarcina species and overproducing several (small ORF encoded) small proteins suggests key physiological functions. Overall, the comprehensive analysis opens an avenue to elucidate the function(s) of multitudinous small proteins and dual-function RNAs in M. mazei.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

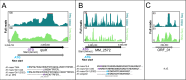

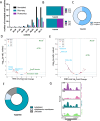
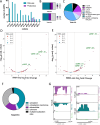

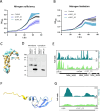
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- RSCHM1052/20-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- RSCHM1052/20-2/Deutsche Forschungsgemeinschaft (German Research Foundation)
- RSCHM1052/19-2/Deutsche Forschungsgemeinschaft (German Research Foundation)
- CIBSS-EXC-2189-390939984/Deutsche Forschungsgemeinschaft (German Research Foundation)
- BA2168/21-2/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

