Cryo-EM analysis of Pseudomonas phage Pa193 structural components
- PMID: 39370451
- PMCID: PMC11456595
- DOI: 10.1038/s42003-024-06985-x
Cryo-EM analysis of Pseudomonas phage Pa193 structural components
Abstract
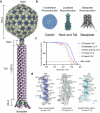
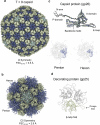
The World Health Organization has designated Pseudomonas aeruginosa as a critical pathogen for the development of new antimicrobials. Bacterial viruses, or bacteriophages, have been used in various clinical settings, commonly called phage therapy, to address this growing public health crisis. Here, we describe a high-resolution structural atlas of a therapeutic, contractile-tailed Pseudomonas phage, Pa193. We used bioinformatics, proteomics, and cryogenic electron microscopy single particle analysis to identify, annotate, and build atomic models for 21 distinct structural polypeptide chains forming the icosahedral capsid, neck, contractile tail, and baseplate. We identified a putative scaffolding protein stabilizing the interior of the capsid 5-fold vertex. We also visualized a large portion of Pa193 ~ 500 Å long tail fibers and resolved the interface between the baseplate and tail fibers. The work presented here provides a framework to support a better understanding of phages as biomedicines for phage therapy and inform engineering opportunities.
© 2024. The Author(s).
Conflict of interest statement
J.R., E.S., R.G., A.S., L.S., P.K., D.B., and S.L. are employees of Armata Pharmaceuticals Inc., a company involved in the development of bacteriophage therapies. LG and JW were contracted by Armata under a fee-for-service agreement. The other authors declare that the research was conducted in a way that is free of financial or commercial relationship that could be construed as conflict of interest.
Figures








Update of
-
Cryo-EM analysis of Pseudomonas phage Pa193 structural components.Res Sq [Preprint]. 2024 Apr 12:rs.3.rs-4189479. doi: 10.21203/rs.3.rs-4189479/v1. Res Sq. 2024. Update in: Commun Biol. 2024 Oct 6;7(1):1275. doi: 10.1038/s42003-024-06985-x. PMID: 38659960 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- S10 OD030457/OD/NIH HHS/United States
- R35 GM140733/GM/NIGMS NIH HHS/United States
- R35GM140733/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R21 AI121531/AI/NIAID NIH HHS/United States
- U24GM129547/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
LinkOut - more resources
Full Text Sources

