Exploring the dynamics and interactions of the N-myc transactivation domain through solution nuclear magnetic resonance spectroscopy
- PMID: 39370942
- PMCID: PMC11555651
- DOI: 10.1042/BCJ20240248
Exploring the dynamics and interactions of the N-myc transactivation domain through solution nuclear magnetic resonance spectroscopy
Abstract
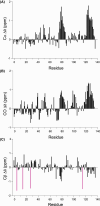
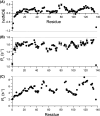
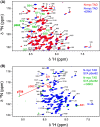
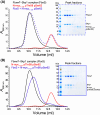
Myc proteins are transcription factors crucial for cell proliferation. They have a C-terminal domain that mediates Max and DNA binding, and an N-terminal disordered region culminating in the transactivation domain (TAD). The TAD participates in many protein-protein interactions, notably with kinases that promote stability (Aurora-A) or degradation (ERK1, GSK3) via the ubiquitin-proteasome system. We probed the structure, dynamics and interactions of N-myc TAD using nuclear magnetic resonance (NMR) spectroscopy following its complete backbone assignment. Chemical shift analysis revealed that N-myc has two regions with clear helical propensity: Trp77-Glu86 and Ala122-Glu132. These regions also have more restricted ps-ns motions than the rest of the TAD, and, along with the phosphodegron, have comparatively high transverse (R2) 15N relaxation rates, indicative of slower timescale dynamics and/or chemical exchange. Collectively these features suggest differential propensities for structure and interaction, either internal or with binding partners, across the TAD. Solution studies on the interaction between N-myc and Aurora-A revealed a previously uncharacterised binding site. The specificity and kinetics of sequential phosphorylation of N-myc by ERK1 and GSK3 were characterised using NMR and resulted in no significant structural changes outside the phosphodegron. When the phosphodegron was doubly phosphorylated, N-myc formed a robust interaction with the Fbxw7-Skp1 complex, but mapping the interaction by NMR suggests a more extensive interface. Our study provides foundational insights into N-myc TAD dynamics and a backbone assignment that will underpin future work on the structure, dynamics, interactions and regulatory post-translational modifications of this key oncoprotein.
Keywords: NMR spectroscopy; intrinsically disordered proteins; myc; neuroblastoma; phosphorylation/dephosphorylation; protein–protein interactions.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures



References
-
- Rickman, D.S., Schulte, J.H. and Eilers, M. (2018) The expanding world of N-MYC–driven tumors. Cancer Discov. 8, 150–163 10.1158/2159-8290.CD-17-0273 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

