This is a preprint.
Biallelic variants in DAP3 result in reduced assembly of the mitoribosomal small subunit with altered intrinsic and extrinsic apoptosis and a Perrault syndrome-spectrum phenotype
- PMID: 39371131
- PMCID: PMC11451657
- DOI: 10.1101/2024.08.19.24312079
Biallelic variants in DAP3 result in reduced assembly of the mitoribosomal small subunit with altered intrinsic and extrinsic apoptosis and a Perrault syndrome-spectrum phenotype
Update in
-
Bi-allelic variants in DAP3 result in reduced assembly of the mitoribosomal small subunit with altered apoptosis and a Perrault-syndrome-spectrum phenotype.Am J Hum Genet. 2025 Jan 2;112(1):59-74. doi: 10.1016/j.ajhg.2024.11.007. Epub 2024 Dec 18. Am J Hum Genet. 2025. PMID: 39701103 Free PMC article.
Abstract
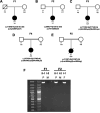
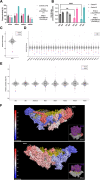
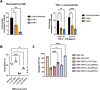
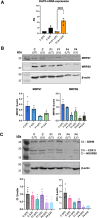
The mitoribosome synthesizes 13 protein subunits of the oxidative phosphorylation system encoded by the mitochondrial genome. The mitoribosome is composed of 12S rRNA, 16S rRNA and 82 mitoribosomal proteins encoded by nuclear genes. To date, variants in 12 genes encoding mitoribosomal proteins are associated with rare monogenic disorders, and frequently show combined oxidative phosphorylation deficiency. Here, we describe five unrelated individuals with biallelic variants in the DAP3 nuclear gene encoding mitoribosomal small subunit 29 (MRPS29), with variable clinical presentations ranging from Perrault syndrome (sensorineural hearing loss and ovarian insufficiency) to an early childhood neurometabolic phenotype. Assessment of respiratory chain function and proteomic profiling of fibroblasts from affected individuals demonstrated reduced MRPS29 protein levels, and consequently decreased levels of additional protein components of the mitoribosomal small subunit, associated with a combined complex I and IV deficiency. Lentiviral transduction of fibroblasts from affected individuals with wild-type DAP3 cDNA increased DAP3 mRNA expression, and partially rescued protein levels of MRPS7, MRPS9 and complex I and IV subunits, demonstrating the pathogenicity of the DAP3 variants. Protein modelling suggested that DAP3 disease-associated missense variants can impact ADP binding, and in vitro assays demonstrated DAP3 variants can consequently reduce both intrinsic and extrinsic apoptotic sensitivity, DAP3 thermal stability and DAP3 GTPase activity. Our study presents genetic and functional evidence that biallelic variants in DAP3 result in a multisystem disorder of combined oxidative phosphorylation deficiency with pleiotropic presentations, consistent with mitochondrial dysfunction.
Keywords: DAP3; MRPS29; Perrault syndrome; leukodystrophy; mitochondria; mitoribosomal small subunit; mitoribosome; ovarian insufficiency; rare disease; sensorineural hearing loss.
Conflict of interest statement
Declaration of interest The authors declare no competing interests.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
