A proteogenomic atlas of the human neural retina
- PMID: 39371417
- PMCID: PMC11450717
- DOI: 10.3389/fgene.2024.1451024
A proteogenomic atlas of the human neural retina
Abstract
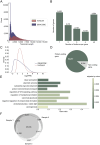
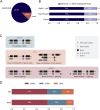
The human neural retina is a complex tissue with abundant alternative splicing and more than 10% of genetic variants linked to inherited retinal diseases (IRDs) alter splicing. Traditional short-read RNA-sequencing methods have been used for understanding retina-specific splicing but have limitations in detailing transcript isoforms. To address this, we generated a proteogenomic atlas that combines PacBio long-read RNA-sequencing data with mass spectrometry and whole genome sequencing data of three healthy human neural retina samples. We identified nearly 60,000 transcript isoforms, of which approximately one-third are novel. Additionally, ten novel peptides confirmed novel transcript isoforms. For instance, we identified a novel IMPDH1 isoform with a novel combination of known exons that is supported by peptide evidence. Our research underscores the potential of in-depth tissue-specific transcriptomic analysis to enhance our grasp of tissue-specific alternative splicing. The data underlying the proteogenomic atlas are available via EGA with identifier EGAD50000000101, via ProteomeXchange with identifier PXD045187, and accessible through the UCSC genome browser.
Keywords: alternative splicing; inherited retinal disease (IRD); isoform; long-read sequencing; mass spectrometry; multi-omics; neural retina; proteogenomics.
Copyright © 2024 Riepe, Stemerdink, Salz, Rey, de Bruijn, Boonen, Tomkiewicz, Kwint, Gloerich, Wessels, Delanote, De Baere, van Nieuwerburgh, De Keulenaer, Ferrari, Ferrari, Coppieters, Cremers, van Wyk, Roosing, de Vrieze and ‘t Hoen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

