Peptide-coated DNA nanostructures as a platform for control of lysosomal function in cells
- PMID: 39372137
- PMCID: PMC11448966
- DOI: 10.1016/j.cej.2024.155633
Peptide-coated DNA nanostructures as a platform for control of lysosomal function in cells
Abstract
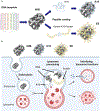
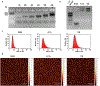
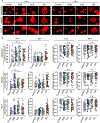
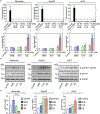
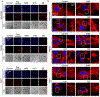
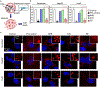
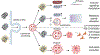
DNA nanotechnology is a rapidly growing field that provides exciting tools for biomedical applications. Targeting lysosomal functions with nanomaterials, such as DNA nanostructures (DNs), represents a rational and systematic way to control cell functionality. Here we present a versatile DNA nanostructure-based platform that can modulate a number of cellular functions depending on the concentration and surface decoration of the nanostructure. Utilizing different peptides for surface functionalization of DNs, we were able to rationally modulate lysosomal activity, which in turn translated into the control of cellular function, ranging from changes in cell morphology to modulation of immune signaling and cell death. Low concentrations of decalysine peptide-coated DNs induced lysosomal acidification, altering the metabolic activity of susceptible cells. In contrast, DNs coated with an aurein-bearing peptide promoted lysosomal alkalization, triggering STING activation. High concentrations of decalysine peptide-coated DNs caused lysosomal swelling, loss of cell-cell contacts, and morphological changes without inducing cell death. Conversely, high concentrations of aurein-coated DNs led to lysosomal rupture and mitochondrial damage, resulting in significant cytotoxicity. Our study holds promise for the rational design of a new generation of versatile DNA-based nanoplatforms that can be used in various biomedical applications, like the development of combinatorial anti-cancer platforms, efficient systems for endolysosomal escape, and nanoplatforms modulating lysosomal pH.
Keywords: DNA nanotechnology; Interferon; Lysosomal rupture; Nanotechnology; bio/nano interactions; lysosome interference.
Figures








References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
