Unveiling genetic links between gut microbiota and asthma: a Mendelian randomization
- PMID: 39372270
- PMCID: PMC11449699
- DOI: 10.3389/fmicb.2024.1448629
Unveiling genetic links between gut microbiota and asthma: a Mendelian randomization
Abstract
Background: Multiple studies suggest a potential connection between the gut microbiome and asthma. Our objective is to use advanced genetic and metagenomic techniques to elucidate the causal relationships and underlying mechanisms between gut microbiota and asthma.
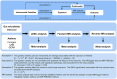
Methods: The study utilized comprehensive Linkage Disequilibrium Score Regression (LDSC) and Mendelian randomization (MR) analyses to examine the relationship between 119 gut microbiota genera and asthma, using publicly accessible genome-wide association studies (GWAS). The meta-analysis synthesized summary effect estimates obtained from LDSC, forward MR, and reverse MR. The MiBioGen collaboration, involving 18,340 individuals, identified genetic variations associated with gut bacteria. Asthma data were collected from the UK Biobank, FinnGen, and GERA, encompassing a total of 82,060 cases and 641,049 controls.
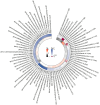
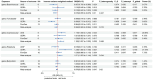
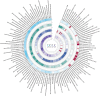
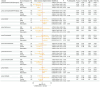
Results: LDSC analysis revealed significant negative genetic correlations between asthma and RuminococcaceaeUCG004 (Rg = -0.55, p = 7.66 × 10-5) and Subdoligranulum (Rg = -0.35, p = 3.61 × 10-4). Forward MR analysis suggested associations between Butyricicoccus (OR = 0.92, p = 0.01), Turicibacter (OR = 0.95, p = 0.025), Butyrivibrio (OR = 0.98, p = 0.047), and reduced asthma risk. Conversely, Coprococcus2 (OR = 1.10, p = 0.035) and Roseburia (OR = 1.07, p = 0.039) were associated with increased risk. Reverse MR analysis indicated significant associations between genetically predicted asthma and Eubacteriumxylanophilumgroup (Beta = -0.08, p = 9.25 × 10-7), LachnospiraceaeNK4A136group (Beta = -0.05, p = 1.26 × 10-4), and Eisenbergiella (Beta = 0.06, p = 0.015, Rg_P = 0.043).
Conclusion: The findings underscore significant genetic correlations and causal relationships between specific gut microbiota and asthma. These insights highlight the potential of gut microbiota as both markers and modulators of asthma risk, offering new avenues for targeted therapeutic strategies.
Keywords: Linkage Disequilibrium Score Regression; Mendelian randomization; asthma; gut microbiota; meta-analysis.
Copyright © 2024 Zheng, Chen, Zhuang, Zhao, Qian and Shi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

