Nitrogen reduces calcium availability by promoting oxalate biosynthesis in apple leaves
- PMID: 39372287
- PMCID: PMC11450213
- DOI: 10.1093/hr/uhae208
Nitrogen reduces calcium availability by promoting oxalate biosynthesis in apple leaves
Abstract
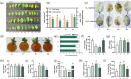
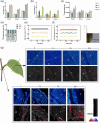
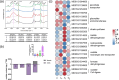
N and Ca are essential nutrients for apple growth and development. Studies have found that Ca content was not low under high N conditions but was poorly available. However, the underlying physiological mechanism through which N regulates Ca availability remains unclear. In this study, apple plants were supplied with N and Ca to analyse the content, in situ distribution, and forms of Ca using noninvasive micro-test technique, electron probe microanalysis, Fourier transform infrared spectroscopy, and transcriptome analysis. A potential interaction was observed between N and Ca in apple leaves. The application of high N and Ca concentration led to a CaOx content of 12.51 g/kg, representing 93.54% of the total Ca in the apple leaves. Electron probe microanalysis revealed that Ca deposited in the phloem primarily existed as CaOx rhombus-shaped crystals. Additionally, high N positively regulated oxalate accumulation in the leaves, increasing it by 40.79 times compared with low N concentration. Specifically, N induced oxalate synthesis in apple leaves by upregulating the MdICL, MdOXAC, and MdMDH genes, while simultaneously inhibiting degradation through downregulation of the MdAAE3 gene. Transcriptome and correlation analyses further confirmed oxaloacetate as the precursor for the synthesis of CaOx crystals in the apple leaves, which were produced via the 'photosynthesis/glycolysis -oxaloacetate -oxalate -CaOx' pathway. WGCNA identified potential regulators of the CaOx biosynthesis pathway triggered by N. Overall, the results provide insights into the regulation of Ca availability by N in apple leaves and support the development of Ca efficient cultivation technique.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Wang WL, Cao ZM, Hou FR. et al. Quality maintenance mechanism of oxalic acid treatment in fresh-cut apple fruit during storage based on nontarget metabolomics analysis. Food Chem. 2024;436:137685. - PubMed
-
- Zhu ZL, Jia ZH, Peng L. et al. Life cycle assessment of conventional and organic apple production systems in China. J Clean Prod. 2018;201:156–68
-
- Freitas STD, Mitcham EJ. Mechanisms regulating apple cultivar susceptibility to bitter pit. Sci Hortic. 2015;186:54–60
LinkOut - more resources
Full Text Sources

