This is a preprint.
Epigenetic control and inheritance of rDNA arrays
- PMID: 39372739
- PMCID: PMC11451732
- DOI: 10.1101/2024.09.13.612795
Epigenetic control and inheritance of rDNA arrays
Abstract
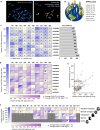
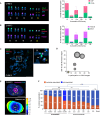
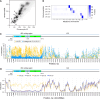
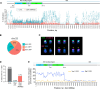
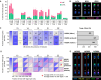
Ribosomal RNA (rRNA) genes exist in multiple copies arranged in tandem arrays known as ribosomal DNA (rDNA). The total number of gene copies is variable, and the mechanisms buffering this copy number variation remain unresolved. We surveyed the number, distribution, and activity of rDNA arrays at the level of individual chromosomes across multiple human and primate genomes. Each individual possessed a unique fingerprint of copy number distribution and activity of rDNA arrays. In some cases, entire rDNA arrays were transcriptionally silent. Silent rDNA arrays showed reduced association with the nucleolus and decreased interchromosomal interactions, indicating that the nucleolar organizer function of rDNA depends on transcriptional activity. Methyl-sequencing of flow-sorted chromosomes, combined with long read sequencing, showed epigenetic modification of rDNA promoter and coding region by DNA methylation. Silent arrays were in a closed chromatin state, as indicated by the accessibility profiles derived from Fiber-seq. Removing DNA methylation restored the transcriptional activity of silent arrays. Array activity status remained stable through the iPS cell re-programming. Family trio analysis demonstrated that the inactive rDNA haplotype can be traced to one of the parental genomes, suggesting that the epigenetic state of rDNA arrays may be heritable. We propose that the dosage of rRNA genes is epigenetically regulated by DNA methylation, and these methylation patterns specify nucleolar organizer function and can propagate transgenerationally.
Figures
References
-
- Bell S. P., Learned R. M., Jantzen H. M. and Tjian R. (1988). “Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis.” Science 241(4870): 1192–1197. - PubMed
-
- Boisvert F. M., van Koningsbruggen S., Navascues J. and Lamond A. I. (2007). “The multifunctional nucleolus.” Nat Rev Mol Cell Biol 8(7): 574–585. - PubMed
-
- Bonduriansky R. (2012). “Rethinking heredity, again.” Trends Ecol Evol 27(6): 330–336. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
