This is a preprint.
Kupffer cell and recruited macrophage heterogeneity orchestrate granuloma maturation and hepatic immunity in visceral leishmaniasis
- PMID: 39372777
- PMCID: PMC11451627
- DOI: 10.1101/2024.07.09.602717
Kupffer cell and recruited macrophage heterogeneity orchestrate granuloma maturation and hepatic immunity in visceral leishmaniasis
Update in
-
Kupffer cell and recruited macrophage heterogeneity orchestrate granuloma maturation and hepatic immunity in visceral leishmaniasis.Nat Commun. 2025 Apr 1;16(1):3125. doi: 10.1038/s41467-025-58360-x. Nat Commun. 2025. PMID: 40169598 Free PMC article.
Abstract
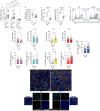
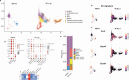
In murine models of visceral leishmaniasis (VL), parasitization of resident Kupffer cells (resKCs) is responsible for early growth of Leishmania infantum in the liver, which leads to granuloma formation and eventual parasite control. We employed the chronic VL model, and revealed an open niche established by KCs death and their migration outside of the sinusoids, resulting in their gradual replacement by monocyte-derived KCs (moKCs). While early granulomas were composed of resKCs, late granulomas were found outside of the sinusoids and contained resKC-derived macrophages, and monocyte-derived macrophages (momacs). ResKCs and moKCs within the sinusoids were identified as homeostatic/regulatory cells, while resKC-derived macrophages and momacs within late granulomas were pro-inflammatory. Despite the infection being largely confined to the resKC-derived macrophages, in the absence of monocyte recruitment, parasite control was strongly compromised. Macrophage heterogeneity, involving migration and reprogramming of resKCs, along with recruitment of monocyte-derived cells, is a hallmark of granuloma maturation and hepatic immunity in VL.
Keywords: Kupffer cells; cell death; chronic infection; ferroptosis; granuloma; liver; monocyte-derived macrophages; parasitic diseases; visceral leishmaniasis.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures









References
-
- Hoeffel G., Chen J., Lavin Y., Low D., Almeida F.F., See P., Beaudin A.E., Lum J., Low I., Forsberg E.C., et al. (2015). C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678. 10.1016/j.immuni.2015.03.011. - DOI - PMC - PubMed
-
- Gomez Perdiguero E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., Garner H., Trouillet C., de Bruijn M.F., Geissmann F., and Rodewald H.R. (2015). Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551. 10.1038/nature13989. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
