This is a preprint.
Characterizing the role of exosomal miRNAs in metastasis
- PMID: 39372783
- PMCID: PMC11451750
- DOI: 10.1101/2024.08.20.608894
Characterizing the role of exosomal miRNAs in metastasis
Update in
-
Characterizing the pan-cancer role of exosomal miRNAs in metastasis across cancers.Comput Struct Biotechnol J. 2024 Dec 26;27:252-264. doi: 10.1016/j.csbj.2024.12.025. eCollection 2025. Comput Struct Biotechnol J. 2024. PMID: 39866667 Free PMC article.
Abstract
Background: Exosomal microRNAs (exomiRs), transported via exosomes, play a pivotal role in intercellular communication. In cancer, exomiRs influence tumor progression by regulating key cellular processes such as proliferation, angiogenesis, and metastasis. Their role in mediating communication between cancer cells and the tumor microenvironment highlights their significance as potential diagnostic and therapeutic targets.
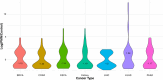
Methodology: In this study, we aimed to characterize the role of exomiRs in influencing the pre-metastatic niche (PMN). Across 7 tumor types, including 4 cell lines and three tumors, we extracted high confidence exomiRs (Log FC >= 2 in exosomes relative to control) and their targets (experimentally identified and targeted by at least 2 exomiRs). Subsequently, we identified enriched pathways and selected the top 100 high-confidence exomiR targets based on the frequency of their appearance in the enriched pathways. These top 100 targets were consistently used throughout the analysis.
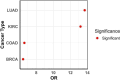
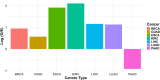
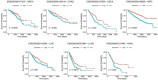
Results: Cancer cell line and tumor derived ExomiRs have significantly higher GC content relative to genomic background. Pathway enriched among the top exomiR targets included general cancer-associated processes such as "wound healing" and "regulation of epithelial cell proliferation", as well as cancer-specific processes, such as "regulation of angiogenesis in kidney" (KIRC), "ossification" in lung (LUAD), and "positive regulation of cytokine production" in pancreatic cancer (PAAD). Similarly, 'Pathways in cancer' and 'MicroRNAs in cancer' ranked among the top 10 enriched KEGG pathways in all cancer types. ExomiR targets were not only enriched for cancer-specific tumor suppressor genes (TSG) but are also downregulated in pre-metastatic niche formed in lungs compared to normal lung. Motif analysis shows high similarity among motifs identified from exomiRs across cancer types. Our analysis recapitulates exomiRs associated with M2 macrophage differentiation and chemoresistance such as miR-21 and miR-222-3p, regulating signaling pathways such as PTEN/PI3/Akt, NF-κB, etc. Cox regression indicated that exomiR targets are significantly associated with overall survival of patients in TCGA. Lastly, a Support Vector Machine (SVM) model using exomiR target gene expression classified responders and non-responders to neoadjuvant chemotherapy with an AUROC of 0.96 (in LUAD), higher than other previously reported gene signatures.
Conclusion: Our study characterizes the pivotal role of exomiRs in shaping the PMN in diverse cancers, underscoring their diagnostic and therapeutic potential.
Keywords: Cancer metastasis; Exosomes; Machine Learning; Survival Analysis; miRNA.
Conflict of interest statement
Declaration of interests I declare no competing interests.
Figures


Similar articles
-
Characterizing the pan-cancer role of exosomal miRNAs in metastasis across cancers.Comput Struct Biotechnol J. 2024 Dec 26;27:252-264. doi: 10.1016/j.csbj.2024.12.025. eCollection 2025. Comput Struct Biotechnol J. 2024. PMID: 39866667 Free PMC article.
-
Prospecting of exosomal-miRNA signatures as prognostic marker for gestational diabetes mellitus and other adverse pregnancy outcomes.Front Endocrinol (Lausanne). 2023 Feb 9;14:1097337. doi: 10.3389/fendo.2023.1097337. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36843574 Free PMC article. Review.
-
Breast Milk-Derived Extracellular Vesicles Enriched in Exosomes From Mothers With Type 1 Diabetes Contain Aberrant Levels of microRNAs.Front Immunol. 2019 Oct 25;10:2543. doi: 10.3389/fimmu.2019.02543. eCollection 2019. Front Immunol. 2019. PMID: 31708933 Free PMC article.
-
Physical Activity as a Preventive Lifestyle Intervention Acts Through Specific Exosomal miRNA Species-Evidence From Human Short- and Long-Term Pilot Studies.Front Physiol. 2021 Aug 2;12:658218. doi: 10.3389/fphys.2021.658218. eCollection 2021. Front Physiol. 2021. PMID: 34408656 Free PMC article.
-
ExomiRs: A Novel Strategy in Cancer Diagnosis and Therapy.Curr Gene Ther. 2018;18(6):336-350. doi: 10.2174/1566523218666181017163204. Curr Gene Ther. 2018. PMID: 30332956 Review.
Cited by
-
Lectin-glycan interactions: a comprehensive cataloguing of cancer-associated glycans for biorecognition and bio-alteration: a review.Glycoconj J. 2024 Oct;41(4-5):301-322. doi: 10.1007/s10719-024-10161-y. Epub 2024 Sep 2. Glycoconj J. 2024. PMID: 39218819 Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
