This is a preprint.
Exploring midgut expression dynamics: longitudinal transcriptomic analysis of adult female Amblyomma americanum midgut and comparative insights with other hard tick species
- PMID: 39372786
- PMCID: PMC11451607
- DOI: 10.1101/2024.09.20.614175
Exploring midgut expression dynamics: longitudinal transcriptomic analysis of adult female Amblyomma americanum midgut and comparative insights with other hard tick species
Abstract
Background: Female ticks remain attached to their host for multiple days to complete a blood meal. This prolonged feeding period is accompanied by a significant increase in the tick's size and body weight, paralleled by noteworthy changes to the tick midgut. While the midgut is recognized for its established role in blood storage and processing, its importance extends to playing a crucial role in the acquisition, survival, and proliferation of pathogens. Despite this, our overall understanding of tick midgut biology is limited.
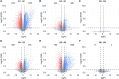
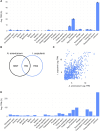
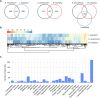
Results: We conducted a comprehensive longitudinal transcriptome analysis of the midgut in adult female A. americanum ticks across various feeding stages, including unfed, slow-feeding, and rapid-feeding phases. Our analysis revealed 15,599 putative DNA coding sequences (CDS) classified within 26 functional groups. Dimensional and differential expression analysis highlighted the dynamic transcriptional changes in the tick midgut as feeding progresses, particularly during the initial period of feeding and the transition from the slow-feeding to the rapid-feeding phase. Additionally, we performed an orthology analysis comparing our dataset with midgut transcriptomes from other hard ticks, such as Ixodes scapularis and Rhipicephalus microplus. This comparison allowed us to identify transcripts commonly expressed during different feeding phases across these three species.
Conclusion: Our findings provide a detailed temporal resolution of numerous metabolic pathways in A. americanum, emphasizing the dynamic transcriptional changes occurring in the tick midgut throughout the feeding process. Furthermore, we identified conserved transcripts across three different tick species that exhibit similar expression patterns. This knowledge has significant implications for future research aimed at deciphering the physiological pathways relevant within the tick midgut. It also offers potential avenues for developing control methods that target multiple tick species.
Keywords: RNA-sequencing; blood meal digestion; midgut; ticks.
Figures


References
-
- Anderson B. E. et al. , Amblyomma americanum: a potential vector of human ehrlichiosis. Am J Trop Med Hyg 49, 239–244 (1993). - PubMed
-
- C. E. a. D. Hopla C.M., The Isolation of Bacterium Tularense from the Tick, Amblyomma Americanum. Journal of the Kansas Entomological Society 26, 2 (1953).
-
- James A. M. et al. , Borrelia lonestari infection after a bite by an Amblyomma americanum tick. J Infect Dis 183, 1810–1814 (2001). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
