This is a preprint.
A Broad Spectrum Lasso Peptide Antibiotic Targeting the Bacterial Ribosome
- PMID: 39372947
- PMCID: PMC11451635
- DOI: 10.21203/rs.3.rs-5058118/v1
A Broad Spectrum Lasso Peptide Antibiotic Targeting the Bacterial Ribosome
Update in
-
A broad-spectrum lasso peptide antibiotic targeting the bacterial ribosome.Nature. 2025 Apr;640(8060):1022-1030. doi: 10.1038/s41586-025-08723-7. Epub 2025 Mar 26. Nature. 2025. PMID: 40140562 Free PMC article.
Abstract
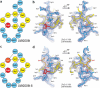
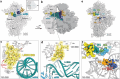
Lasso peptides, biologically active molecules with a distinct structurally constrained knotted fold, are natural products belonging to the class of ribosomally-synthesized and posttranslationally modified peptides (RiPPs). Lasso peptides act upon several bacterial targets, but none have been reported to inhibit the ribosome, one of the main antibiotic targets in the bacterial cell. Here, we report the identification and characterization of the lasso peptide antibiotic, lariocidin (LAR), and its internally cyclized derivative, lariocidin B (LAR-B), produced by Paenabacillussp. M2, with broad-spectrum activity against many bacterial pathogens. We show that lariocidins inhibit bacterial growth by binding to the ribosome and interfering with protein synthesis. Structural, genetic, and biochemical data show that lariocidins bind at a unique site in the small ribosomal subunit, where they interact with the 16S rRNA and aminoacyl-tRNA, inhibiting translocation and inducing miscoding. LAR is unaffected by common resistance mechanisms, has a low propensity for generating spontaneous resistance, shows no human cell toxicity, and has potent in vivo activity in a mouse model of Acinetobacter baumannii infection. Our finding of the first ribosome-targeting lasso peptides uncovers new routes toward discovering alternative protein synthesis inhibitors and offers a new chemical scaffold for developing much-needed antibacterial drugs.
Keywords: Lasso peptide; RiPP; antibiotic; decoding; drug resistance; inhibitor; ribosome; tRNA; translocation.
Conflict of interest statement
COMPETING INTERESTS STATEMENT The authors declare no competing interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

