An Automated Cell-Free Workflow for Transcription Factor Engineering
- PMID: 39373325
- PMCID: PMC11494693
- DOI: 10.1021/acssynbio.4c00471
An Automated Cell-Free Workflow for Transcription Factor Engineering
Abstract
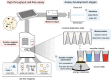
The design and optimization of metabolic pathways, genetic systems, and engineered proteins rely on high-throughput assays to streamline design-build-test-learn cycles. However, assay development is a time-consuming and laborious process. Here, we create a generalizable approach for the tailored optimization of automated cell-free gene expression (CFE)-based workflows, which offers distinct advantages over in vivo assays in reaction flexibility, control, and time to data. Centered around designing highly accurate and precise transfers on the Echo Acoustic Liquid Handler, we introduce pilot assays and validation strategies for each stage of protocol development. We then demonstrate the efficacy of our platform by engineering transcription factor-based biosensors. As a model, we rapidly generate and assay libraries of 127 MerR and 134 CadR transcription factor variants in 3682 unique CFE reactions in less than 48 h to improve limit of detection, selectivity, and dynamic range for mercury and cadmium detection. This was achieved by assessing a panel of ligand conditions for sensitivity (to 0.1, 1, 10 μM Hg and 0, 1, 10, 100 μM Cd for MerR and CadR, respectively) and selectivity (against Ag, As, Cd, Co, Cu, Hg, Ni, Pb, and Zn). We anticipate that our Echo-based, cell-free approach can be used to accelerate multiple design workflows in synthetic biology.
Keywords: cell-free gene expression; high-throughput; protein engineering; robotic liquid handling; synthetic biology; transcription factor.
Conflict of interest statement
The authors declare the following competing financial interest(s): M.C.J. is a co-founder and has financial interest in Stemloop, Inc., Pearl Bio, Gauntlet Bio, and Synolo Therapeutics; J.B.L. is a co-founder and has financial interest in Stemloop. Inc. These interests are reviewed and managed by Northwestern University and Stanford University in accordance with their conflict-of-interest policies. All other authors report no competing interests.
Figures



References
-
- Garenne D.; Haines M. C.; Romantseva E. F.; Freemont P.; Strychalski E. A.; Noireaux V. Cell-free gene expression. Nat. Rev. Methods Primer 2021, 1, 49.10.1038/s43586-021-00046-x. - DOI
-
- Martin R. W.; Des Soye B. J.; Kwon Y.-C.; Kay J.; Davis R. G.; Thomas P. M.; Majewska N. I.; Chen C. X.; Marcum R. D.; Weiss M. G.; Stoddart A. E.; Amiram M.; Ranji Charna A. K.; Patel J. R.; Isaacs F. J.; Kelleher N. L.; Hong S. H.; Jewett M. C. Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids. Nat. Commun. 2018, 9, 1203.10.1038/s41467-018-03469-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

