The development of single-domain VHH nanobodies that target the Candida albicans cell surface
- PMID: 39373478
- PMCID: PMC11572700
- DOI: 10.1128/spectrum.04269-23
The development of single-domain VHH nanobodies that target the Candida albicans cell surface
Abstract
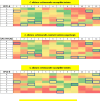
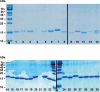
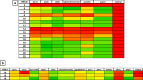
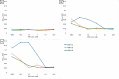
Candida albicans causes life-threatening invasive infections that are hard to diagnose and treat, with drug resistance leading to treatment failure. The goal of this study was to develop VHH (single variable domain on a heavy chain) nanobodies to detect drug-resistant infections. Llamas were immunized with a mixture of heat killed and fixed C. albicans cells of different morphologies. Llama lymphocyte RNA was used to generate phage display libraries that were tested for binding to C. albicans cells or cell wall fractions, and single antibody domains were isolated. The libraries were panned against echinocandin-resistant C. albicans isolates and counter-selected against echinocandin-susceptible isolates with the aim of isolating binding domains specific for antigens on drug-resistant cells. Thirty diverse VHH nanobodies were selected, and binding characteristics were assessed via dose-response ELISA. Binding was tested against a variety of C. albicans isolates and other Candida species, indicating that the VHHs were specific for C. albicans. The VHH nanobodies were sorted into four distinct groups based on their binding patterns. Two of the groups bound preferentially to the yeast cell poles and hyphae, respectively. Nanobody binding to C. albicans deletion mutants was tested by fluorescence microscopy and ELISA to identify the antigen targets. VHH19 nanobody, belonging to the largest group, recognized the Als4 adhesin. VHH14 antibody in the hyphae-specific group recognized Als3. None of the isolated VHH nanobodies was selective for drug-resistant clinical isolates. Our data indicate that this approach can generate valuable single-domain antibodies specific to C. albicans proteins.IMPORTANCEThe human fungal pathogen Candida albicans causes a range of diseases from superficial mucosal infections such as oral and vaginal thrush to life-threatening, systemic infections. Accurate and rapid diagnosis of these infections remains challenging, and currently, there are no rapid ways to diagnose drug-resistant infections without performing drug susceptibility testing from blood culture, which can take several days. In this proof-of-concept study, we have generated a diverse set of single domain VHH antibodies (nanobodies) from llamas that recognize and bind specifically to C. albicans cell surface. The nanobodies were classified into four groups based on their binding patterns, for example, cell poles or hyphae. Specific nanobodies were verified as recognizing the important adhesin Als4 or the hyphae associated invasin Als3, respectively. The data validate the approach that small VHH antibody domains hold future promise for diagnostic applications and as probes to study the fungal cell surface.
Keywords: Candida albicans; antibodies; fungal cell wall.
Conflict of interest statement
E.D. and T.C.V. are affiliated with QVQ company.
Figures













References
-
- Fisher MC, Alastruey-Izquierdo A, Berman J, Bicanic T, Bignell EM, Bowyer P, Bromley M, Brüggemann R, Garber G, Cornely OA, Gurr SJ, Harrison TS, Kuijper E, Rhodes J, Sheppard DC, Warris A, White PL, Xu J, Zwaan B, Verweij PE. 2022. Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol 20:557–571. doi: 10.1038/s41579-022-00720-1 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

