oriTDB: a database of the origin-of-transfer regions of bacterial mobile genetic elements
- PMID: 39373502
- PMCID: PMC11701681
- DOI: 10.1093/nar/gkae869
oriTDB: a database of the origin-of-transfer regions of bacterial mobile genetic elements
Abstract
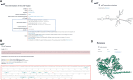
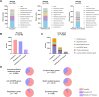
Conjugation and mobilization are two important pathways of horizontal transfer of bacterial mobile genetic elements (MGEs). The origin-of-transfer (oriT) region is crucial for this process, serving as a recognition site for relaxase and containing the DNA nicking site (nic site), which initiates the conjugation or mobilization. Here, we present a database of the origin-of-transfer regions of bacterial MGEs, oriTDB (https://bioinfo-mml.sjtu.edu.cn/oriTDB2/). Incorporating data from text mining and genome analysis, oriTDB comprises 122 experimentally validated and 22 927 predicted oriTs within bacterial plasmids, Integrative and Conjugative Elements, and Integrative and Mobilizable Elements. Additionally, oriTDB includes details about associated relaxases, auxiliary proteins, type IV coupling proteins, and a gene cluster encoding the type IV secretion system. The database also provides predicted secondary structures of oriT sequences, dissects oriT regions into pairs of inverted repeats, nic sites, and their flanking conserved sequences, and offers an interactive visual representation. Furthermore, oriTDB includes an enhanced oriT prediction pipeline, oriTfinder2, which integrates a functional annotation module for cargo genes in bacterial MGEs. This resource is intended to support research on bacterial conjugative or mobilizable elements and promote an understanding of their cargo gene functions.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Frost L.S., Leplae R., Summers A.O., Toussaint A.. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Micro. 2005; 3:722–732. - PubMed
-
- de la Cruz F., Frost L.S., Meyer R.J., Zechner E.L. Conjugative DNA metabolism in gram-negative bacteria. FEMS Microbiol. Rev. 2010; 34:18–40. - PubMed
-
- Garcillán-Barcia M.P., Redondo-Salvo S., de la Cruz F.. Plasmid classifications. Plasmid. 2023; 126:102684. - PubMed
-
- Francia M.V., Varsaki A., Garcillán-Barcia M.P., Latorre A., Drainas C., de la Cruz F.. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 2004; 28:79–100. - PubMed
MeSH terms
Substances
Grants and funding
- 2023YFC2307103/National Key Research and Development Program of China
- 32070572/National Natural Science Foundation of China
- SHDC2022CRD039/Shanghai Hospital Development Center Foundation
- GWV1-11.2-XD01/Talent Program on Public Health System Construction of Shanghai
- 20240818/Shanghai Jiao Tong University School of Medicine
LinkOut - more resources
Full Text Sources

