The HSV-1 encoded CCCTC-binding factor, CTRL2, impacts the nature of viral chromatin during HSV-1 lytic infection
- PMID: 39374265
- PMCID: PMC11486355
- DOI: 10.1371/journal.ppat.1012621
The HSV-1 encoded CCCTC-binding factor, CTRL2, impacts the nature of viral chromatin during HSV-1 lytic infection
Abstract
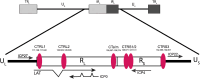
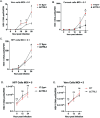
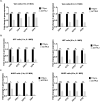
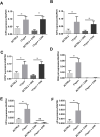
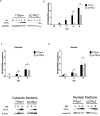
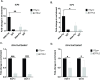
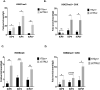
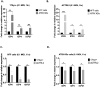
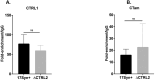
HSV-1 genomes are rapidly heterochromatinized following entry by host cells to limit viral gene expression. Efficient HSV-1 genome replication requires mechanisms that de-repress chromatin associated with the viral genome. CCCTC-binding factors, or CTCF insulators play both silencing and activating roles in cellular transcriptional regulation. Importantly, the HSV-1 genome encodes several CTCF insulators that flank IE genes, implying that individual HSV-1 encoded CTCF insulators regulate IE transcription during all stages of the HSV-1 life cycle. We previously reported that the HSV-1 encoded CTCF insulator located downstream of the LAT (CTRL2) controlled IE gene silencing during latency. To further characterize the role of this insulator during the lytic infection we leveraged a ΔCTRL2 recombinant virus to show that there was a genome replication defect that stemmed from decreased IE gene expression in fibroblasts and epithelial cells at early times following initiation of infection. Further experiments indicated that the defect in gene expression resulted from chromatin inaccessibility in the absence of the insulator. To elucidate how chromatin accessibility was altered in the absence of the CTRL2 insulator, we showed that enrichment of Alpha-thalassemia/mental retardation, X-linked chromatin remodeler (ATRX), and the histone variant H3.3, both of which are known for their roles in maintaining repressive histone markers on the HSV-1 viral genome were increased on IE regions of HSV-1. Finally, both H3K27me3 and H3K9me3 repressive histone marks remained enriched by 4 hours post infection in the absence of the CTRL2 insulator, confirming that the CTRL2 insulator is required for de-repression of IE genes of viral genomes. To our knowledge these are the first data that show that a specific CTCF insulator in the HSV-1 genome (CTRL2) regulates chromatin accessibility during the lytic infection.
Copyright: © 2024 Singh et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Neumann DM, Bhattacharjee PS, Giordani NV, Bloom DC, Hill JM. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J Virol. 2007;81(23):13248–53. Epub 20070919. doi: 10.1128/JVI.01569-07 ; PubMed Central PMCID: PMC2169074. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

