Impaired axon initial segment structure and function in a model of ARHGEF9 developmental and epileptic encephalopathy
- PMID: 39374387
- PMCID: PMC11494352
- DOI: 10.1073/pnas.2400709121
Impaired axon initial segment structure and function in a model of ARHGEF9 developmental and epileptic encephalopathy
Abstract
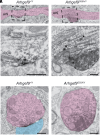
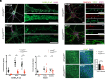
Developmental and epileptic encephalopathies (DEE) are rare but devastating and largely intractable childhood epilepsies. Genetic variants in ARHGEF9, encoding a scaffolding protein important for the organization of the postsynaptic density of inhibitory synapses, are associated with DEE accompanied by complex neurological phenotypes. In a mouse model carrying a patient-derived ARHGEF9 variant associated with severe disease, we observed aggregation of postsynaptic proteins and loss of functional inhibitory synapses at the axon initial segment (AIS), altered axo-axonic synaptic inhibition, disrupted action potential generation, and complex seizure phenotypes consistent with clinical observations. These results illustrate diverse roles of ARHGEF9 that converge on regulation of the structure and function of the AIS, thus revealing a pathological mechanism for ARHGEF9-associated DEE. This unique example of a neuropathological condition associated with multiple AIS dysfunctions may inform strategies for treating neurodevelopmental diseases.
Keywords: axon initial segment; epilepsy; hippocampus; mouse model.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




References
-
- Kins S., Betz H., Kirsch J., Collybistin, a newly identified brain-specific GEF, induces submembrane clustering of gephyrin. Nat. Neurosci. 3, 22–29 (2000). - PubMed
-
- Poulopoulos A., et al. , Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron 63, 628–642 (2009). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

