Upregulation of nuclear protein Hemgn by transcriptional repressor Gfi1 through repressing PU.1 contributes to the anti-apoptotic activity of Gfi1
- PMID: 39374784
- PMCID: PMC11550643
- DOI: 10.1016/j.jbc.2024.107860
Upregulation of nuclear protein Hemgn by transcriptional repressor Gfi1 through repressing PU.1 contributes to the anti-apoptotic activity of Gfi1
Abstract
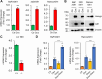
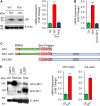
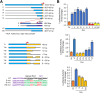
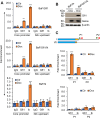
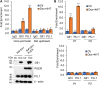
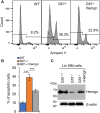
Gfi1 is a transcriptional repressor that plays a critical role in hematopoiesis. The repressive activity of Gfi1 is mediated mainly by its SNAG domain that interacts with and thereby recruits the histone demethylase LSD1 to its target genes. An important function of Gfi1 is to protect hematopoietic cells against stress-induced apoptosis, which has been attributed to its participation in the posttranscriptional modifications of p53 protein, leading to suppression of p53 activity. In this study, we show that Gfi1 upregulated the expression of Hemgn, a nuclear protein, through a 16-bp promoter region spanning from +47 to +63 bp relative to the transcription start site (TSS), which was dependent on its interaction with LSD1. We further demonstrate that Gfi1, Ikaros, and PU.1 are bound to this 16-bp region. However, while Ikaros activated Hemgn and collaborated with Gfi1 to augment Hemgn expression, it was not required for Gfi1-mediated Hemgn upregulation. In contrast, PU.1 repressed Hemgn and inhibited Hemgn upregulation by Gfi1. Notably, PU.1 knockdown and deficiency, while augmenting Hemgn expression, abolished Hemgn upregulation by Gfi1. PU.1 (Spi-1) is repressed by Gfi1. We show here that PU.1 repression by Gfi1 preceded and correlated well with Hemgn upregulation. Thus, our data strongly suggests that Gfi1 upregulates Hemgn by repressing PU.1. In addition, we demonstrate that Hemgn upregulation contributed to the anti-apoptotic activity of Gfi1 in a p53-independent manner.
Keywords: Gfi1; Hemgn; Ikaros; PU.1; apoptosis; cell death; histone demethylase; transcription; transcription repressor.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
-
- Hock H., Hamblen M.J., Rooke H.M., Schindler J.W., Saleque S., Fujiwara Y., et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. - PubMed
-
- Khandanpour C., Kosan C., Gaudreau M.C., Dührsen U., Hébert J., Zeng H., et al. Growth factor independence 1 protects hematopoietic stem cells against apoptosis but also prevents the development of a myeloproliferative-like disease. Stem Cells. 2011;29:376–385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

