HNRNPC mediates lymphatic metastasis of cervical cancer through m6A-dependent alternative splicing of FOXM1
- PMID: 39375330
- PMCID: PMC11458786
- DOI: 10.1038/s41419-024-07108-4
HNRNPC mediates lymphatic metastasis of cervical cancer through m6A-dependent alternative splicing of FOXM1
Abstract
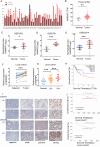
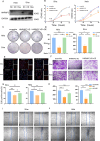
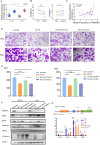
Cervical cancer (CCa) patients with lymph node (LN) metastasis face poor prognoses and have limited treatment options. Aberrant N6-methyladenosine (m6A) modification of RNAs are known to promote tumor metastasis, but their role in CCa remains unclear. Our study reveals that HNRNPC, an alternative splicing (AS) factor and m6A reader, increases tumor-related variants through m6A-dependent manner, thereby promoting lymphatic metastasis in CCa. We found that HNRNPC overexpression correlates with lymphatic metastasis and poorer prognoses in CCa patients. Functionally, knocking down HNRNPC markedly inhibited the migration and invasion of several CCa cell lines, while supplementing HNRNPC restored the malignant phenotypes of these cells. Mechanistically, HNRNPC regulates exon skipping of FOXM1 by binding to its m6A-modified motif. Mutating the m6A site on FOXM1 weakened the interaction between HNRNPC and FOXM1 pre-RNA, leading to a reduction in the metastasis-related FOXM1-S variant. In conclusion, our findings demonstrate that m6A-dependent alternative splicing mediated by HNRNPC is essential for lymphatic metastasis in CCa, potentially providing novel clinical markers and therapeutic strategies for patients with advanced CCa.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

