Structure of the Nipah virus polymerase phosphoprotein complex
- PMID: 39375338
- PMCID: PMC11458586
- DOI: 10.1038/s41467-024-52701-y
Structure of the Nipah virus polymerase phosphoprotein complex
Abstract
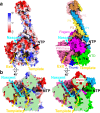
The Nipah virus (NiV), a member of the Paramyxoviridae family, is notorious for its high fatality rate in humans. The RNA polymerase machinery of NiV, comprising the large protein L and the phosphoprotein P, is essential for viral replication. This study presents the 2.9-Å cryo-electron microscopy structure of the NiV L-P complex, shedding light on its assembly and functionality. The structure not only demonstrates the molecular details of the conserved N-terminal domain, RNA-dependent RNA polymerase (RdRp), and GDP polyribonucleotidyltransferase of the L protein, but also the intact central oligomerization domain and the C-terminal X domain of the P protein. The P protein interacts extensively with the L protein, forming an antiparallel β-sheet among the P protomers and with the fingers subdomain of RdRp. The flexible linker domain of one P promoter extends its contact with the fingers subdomain to reach near the nascent RNA exit, highlighting the distinct characteristic of the NiV L-P interface. This distinctive tetrameric organization of the P protein and its interaction with the L protein provide crucial molecular insights into the replication and transcription mechanisms of NiV polymerase, ultimately contributing to the development of effective treatments and preventive measures against this Paramyxoviridae family deadly pathogen.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Chua, K. B. et al. Nipah virus: a recently emergent deadly paramyxovirus. Science288, 1432–1435 (2000). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

