The activating receptor NKG2D is an anti-fungal pattern recognition receptor
- PMID: 39375344
- PMCID: PMC11458907
- DOI: 10.1038/s41467-024-52913-2
The activating receptor NKG2D is an anti-fungal pattern recognition receptor
Abstract
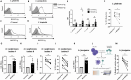
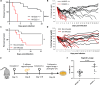
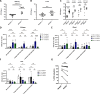
NKG2D is a central activating receptor involved in target recognition and killing by Natural Killer and CD8+ T cells. The known role of NKG2D is to recognize a family of self-induced stress ligands that are upregulated on stressed cells such as cancerous or virally infected cells. Fungal pathogens are a major threat to human health, infecting more than a billion patients yearly and becoming more common and drug resistant. Here we show that NKG2D plays a critical role in the immune response against fungal infections. NKG2D can recognize fungal pathogens from most major families including Candida, Cryptococcus and Aspergillus species, and mice lacking NKG2D are extremely sensitive to fungal infections in models of both invasive and mucosal infections, making NKG2D an anti-fungal pattern recognition receptor.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

