Glycerophospholipid remodeling is critical for orthoflavivirus infection
- PMID: 39375358
- PMCID: PMC11458896
- DOI: 10.1038/s41467-024-52979-y
Glycerophospholipid remodeling is critical for orthoflavivirus infection
Abstract
Flavivirus infection is tightly connected to host lipid metabolism. Here, we performed shotgun lipidomics of cells infected with neurotropic Zika, West Nile, and tick-borne encephalitis virus, as well as dengue and yellow fever virus. Early in infection specific lipids accumulate, e.g., neutral lipids in Zika and some lysophospholipids in all infections. Ceramide levels increase following infection with viruses that cause a cytopathic effect. In addition, fatty acid desaturation as well as glycerophospholipid metabolism are significantly altered. Importantly, depletion of enzymes involved in phosphatidylserine metabolism as well as phosphatidylinositol biosynthesis reduce orthoflavivirus titers and cytopathic effects while inhibition of fatty acid monounsaturation only rescues from virus-induced cell death. Interestingly, interfering with ceramide synthesis has opposing effects on virus replication and cytotoxicity depending on the targeted enzyme. Thus, lipid remodeling by orthoflaviviruses includes distinct changes but also common patterns shared by several viruses that are needed for efficient infection and replication.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
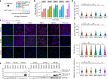




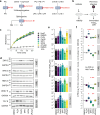

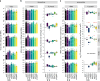

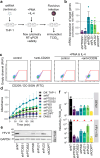
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 517270163/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 517270083/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 197785619 (CRC 2021 project B10)/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 416701689/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 197785619 (CRC 2021 project Z02)/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

