GeneCompass: deciphering universal gene regulatory mechanisms with a knowledge-informed cross-species foundation model
- PMID: 39375485
- PMCID: PMC11615217
- DOI: 10.1038/s41422-024-01034-y
GeneCompass: deciphering universal gene regulatory mechanisms with a knowledge-informed cross-species foundation model
Abstract
Deciphering universal gene regulatory mechanisms in diverse organisms holds great potential for advancing our knowledge of fundamental life processes and facilitating clinical applications. However, the traditional research paradigm primarily focuses on individual model organisms and does not integrate various cell types across species. Recent breakthroughs in single-cell sequencing and deep learning techniques present an unprecedented opportunity to address this challenge. In this study, we built an extensive dataset of over 120 million human and mouse single-cell transcriptomes. After data preprocessing, we obtained 101,768,420 single-cell transcriptomes and developed a knowledge-informed cross-species foundation model, named GeneCompass. During pre-training, GeneCompass effectively integrated four types of prior biological knowledge to enhance our understanding of gene regulatory mechanisms in a self-supervised manner. By fine-tuning for multiple downstream tasks, GeneCompass outperformed state-of-the-art models in diverse applications for a single species and unlocked new realms of cross-species biological investigations. We also employed GeneCompass to search for key factors associated with cell fate transition and showed that the predicted candidate genes could successfully induce the differentiation of human embryonic stem cells into the gonadal fate. Overall, GeneCompass demonstrates the advantages of using artificial intelligence technology to decipher universal gene regulatory mechanisms and shows tremendous potential for accelerating the discovery of critical cell fate regulators and candidate drug targets.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

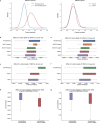
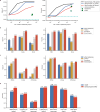
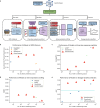
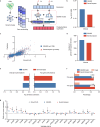

References
MeSH terms
Grants and funding
- No.31971289/National Natural Science Foundation of China (National Science Foundation of China)
- No.91954201/National Natural Science Foundation of China (National Science Foundation of China)
- No.62202455/National Natural Science Foundation of China (National Science Foundation of China)
- No.32341013/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Other Literature Sources

