Identifying patient subgroups in MASLD and MASH-associated fibrosis: molecular profiles and implications for drug development
- PMID: 39375498
- PMCID: PMC11458909
- DOI: 10.1038/s41598-024-74098-w
Identifying patient subgroups in MASLD and MASH-associated fibrosis: molecular profiles and implications for drug development
Abstract
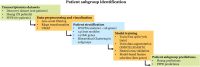
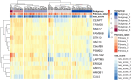
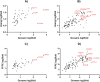
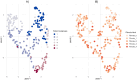
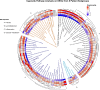
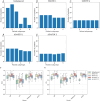
The incidence of MASLD and MASH-associated fibrosis is rapidly increasing worldwide. Drug therapy is hampered by large patient variability and partial representation of human MASH fibrosis in preclinical models. Here, we investigated the mechanisms underlying patient heterogeneity using a discovery dataset and validated in distinct human transcriptomic datasets, to improve patient stratification and translation into subgroup specific patterns. Patient stratification was performed using weighted gene co-expression network analysis (WGCNA) in a large public transcriptomic discovery dataset (n = 216). Differential expression analysis was performed using DESeq2 to obtain differentially expressed genes (DEGs). Ingenuity Pathway analysis was used for functional annotation. The discovery dataset showed relevant fibrosis-related mechanisms representative of disease heterogeneity. Biological complexity embedded in genes signature was used to stratify discovery dataset into six subgroups of various sizes. Of note, subgroup-specific DEGs show differences in directionality in canonical pathways (e.g. Collagen biosynthesis, cytokine signaling) across subgroups. Finally, a multiclass classification model was trained and validated in two datasets. In summary, our work shows a potential alternative for patient population stratification based on heterogeneity in MASLD-MASH mechanisms. Future research is warranted to further characterize patient subgroups and identify protein targets for virtual screening and/or in vitro validation in preclinical models.
Keywords: Biological patterns; Heterogeneity; Individual variation; Liver disease; Patient stratification; Subgroup-specific pathways.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

