Differential gene expression in neonatal calf muscle tissues from Hanwoo cows overfed during mid to late pregnancy period
- PMID: 39375502
- PMCID: PMC11458785
- DOI: 10.1038/s41598-024-74976-3
Differential gene expression in neonatal calf muscle tissues from Hanwoo cows overfed during mid to late pregnancy period
Abstract
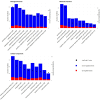
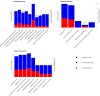
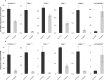
Maternal nutrition significantly influences fetal development and postnatal outcomes. This study investigates the impact of maternal overfeeding during mid to late pregnancy on gene expression in the round and sirloin muscles of Hanwoo neonatal calves. Eight cows were assigned to either a control group receiving standard nutrition (100%) or a treated group receiving overnutrition (150%). After birth, tissue samples from the round and sirloin muscles of neonatal calves were collected and subjected to RNA sequencing to assess differentially expressed genes (DEGs). RNA sequencing identified 43 DEGs in round muscle and 15 in sirloin muscle, involving genes related to myogenesis, adipogenesis, and energy regulation. Key genes, including PPARGC1A, THBS1, CD44, JUND, CNN1, ENAH, and RUNX1, were predominantly downregulated. Gene ontology (GO) enrichment analyses revealed terms associated with muscle development, such as "biological regulation," "cellular process," and "response to stimulus." Protein-protein interaction networks highlighted complex interactions among DEGs. Random Forest analysis identified ARC, SLC1A5, and GNPTAB as influential genes for distinguishing between control and treated groups. Overall, maternal overnutrition during mid-to-late pregnancy results in the downregulation of genes involved in muscle development and energy metabolism in neonatal Hanwoo calves. These findings provide insights into the molecular effects of maternal nutrition on muscle development.
Keywords: Beef cattle metabolism; Maternal nutrition; Metabolic programming; Skeletal muscle development; Transcriptomic analysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Noya, A., Ripoll, G., Casasús, I. & Sanz, A. Long-term effects of early maternal undernutrition on the growth, physiological profiles, carcass and meat quality of male beef offspring. Res. Vet. Sci.142, 1–11 (2022). - PubMed
-
- Marquez, D. et al. Supplementation of grazing beef cows during gestation as a strategy to improve skeletal muscle development of the offspring. Anim. 11, 2184–2192 (2017). - PubMed
-
- Du, M., Ford, S. P. & Zhu, M. J. Optimizing livestock production efficiency through maternal nutritional management and fetal developmental programming. Anim. Front.7, 5–11 (2017).
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

