Alternative splicing of PBRM1 mediates resistance to PD-1 blockade therapy in renal cancer
- PMID: 39375538
- PMCID: PMC11574163
- DOI: 10.1038/s44318-024-00262-7
Alternative splicing of PBRM1 mediates resistance to PD-1 blockade therapy in renal cancer
Abstract
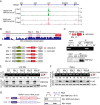
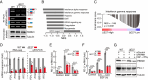
Alternative pre-mRNA splicing (AS) is a biological process that results in proteomic diversity. However, implications of AS alterations in cancer remain poorly understood. Herein, we performed a comprehensive AS analysis in cancer driver gene transcripts across fifteen cancer types and found global alterations in inclusion rates of the PBAF SWI/SNF chromatin remodeling complex subunit Polybromo 1 (PBRM1) exon 27 (E27) in most types of cancer tissues compared with those in normal tissues. Further analysis confirmed that PBRM1 E27 is excluded by the direct binding of RBFOX2 to intronic UGCAUG elements. In addition, the E27-included PBRM1 isoform upregulated PD-L1 expression via enhanced PBAF complex recruitment to the PD-L1 promoter. PBRM1 wild-type patients with clear cell renal cell carcinoma were resistant to PD-1 blockade therapy when they expressed low RBFOX2 mRNA levels. Overall, our study suggests targeting of RBFOX2-mediated AS of PBRM1 as a potential therapeutic strategy for immune checkpoint blockade.
Keywords: Alternative Splicing; Immune Checkpoint Inhibitor; PBRM1; PD-L1; RBFOX2.
© 2024. The Author(s).
Conflict of interest statement
Figures









References
-
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R et al (2012) The evolutionary landscape of alternative splicing in vertebrate species. Science 338:1587–1593 - PubMed
-
- Bludau I, Aebersold R (2020) Proteomic and interactomic insights into the molecular basis of cell functional diversity. Nat Rev Mol Cell Bio 21:327–340 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

