Genetic architecture reconciles linkage and association studies of complex traits
- PMID: 39375568
- PMCID: PMC11835202
- DOI: 10.1038/s41588-024-01940-2
Genetic architecture reconciles linkage and association studies of complex traits
Abstract
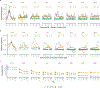
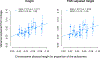
Linkage studies have successfully mapped loci underlying monogenic disorders, but mostly failed when applied to common diseases. Conversely, genome-wide association studies (GWASs) have identified replicable associations between thousands of SNPs and complex traits, yet capture less than half of the total heritability. In the present study we reconcile these two approaches by showing that linkage signals of height and body mass index (BMI) from 119,000 sibling pairs colocalize with GWAS-identified loci. Concordant with polygenicity, we observed the following: a genome-wide inflation of linkage test statistics; that GWAS results predict linkage signals; and that adjusting phenotypes for polygenic scores reduces linkage signals. Finally, we developed a method using recombination rate-stratified, identity-by-descent sharing between siblings to unbiasedly estimate heritability of height (0.76 ± 0.05) and BMI (0.55 ± 0.07). Our results imply that substantial heritability remains unaccounted for by GWAS-identified loci and this residual genetic variation is polygenic and enriched near these loci.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
COMPETING INTERESTS
The authors declare no competing interests.
Figures

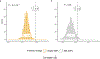
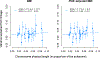



References
-
- Polderman TJC et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47, 702–9 (2015). - PubMed
-
- Risch N & Merikangas K The future of genetic studies of complex human diseases. Science 273, 1516–1517 (1996). - PubMed
-
- Lynch M & Walsh B Genetics and Analysis of Quantitative Traits. (Sinauer Associates, Inc., Sunderland, MA, 1998).
-
- Botstein D & Risch N Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet 33, 228–237 (2003). - PubMed
-
- Hall JM et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250, 1684–1689 (1990). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

