Genomic and transcriptomic profiling of pre- and postneoadjuvant chemotherapy triple negative breast cancer tumors
- PMID: 39375938
- PMCID: PMC11611771
- DOI: 10.1111/cas.16339
Genomic and transcriptomic profiling of pre- and postneoadjuvant chemotherapy triple negative breast cancer tumors
Abstract
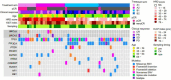
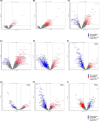
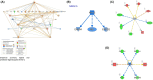
Our understanding of neoadjuvant treatment with microtubule inhibitors (MTIs) for triple negative breast cancer (TNBC) remains limited. To advance our understanding of the role of breast cancer driver genes' mutational status with pathological complete response (pCR; ypT0/isypN0) prediction and to identify distinct gene sets for MTIs like eribulin and paclitaxel, we carried out targeted genomic (n = 50) and whole transcriptomic profiling (n = 64) of TNBC tumor samples from the Japan Breast Cancer Research Group 22 (JBCRG-22) clinical trial. Lower PIK3CA, PTEN, and HRAS mutations were found in homologous recombination deficiency (HRD)-high (HRD score ≥ 42) tumors with higher pCR rates. When HRD-high tumors were stratified by tumor BRCA mutation status, the pCR rates in BRCA2-mutated tumors were higher (83% vs. 36%). Transcriptomic profiling of TP53-positive tumors identified downregulation of FGFR2 (false discovery rate p value = 2.07e-7), which was also the only common gene between HRD-high and -low tumors with pCR/quasi-pCR treated with paclitaxel and eribulin combined with carboplatin, respectively. Differential enrichment analysis of the HRD-high group posttreatment tumors revealed significant correlation (p = 0.006) of the glycan degradation pathway. FGFR2 expression and the differentially enriched pathways play a role in the response and resistance to MTIs containing carboplatin treatment in TNBC patients.
Keywords: FGFR2; HRD; eribulin; microtubule inhibitor; triple negative breast cancer.
© 2024 The Author(s). Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
SO, SOg, and MT are editorial board members of
Figures
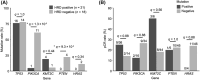


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

