The involvement of multiple ABC transporters in daunorubicin efflux in Streptomyces coeruleorubidus
- PMID: 39375957
- PMCID: PMC11458662
- DOI: 10.1111/1751-7915.70023
The involvement of multiple ABC transporters in daunorubicin efflux in Streptomyces coeruleorubidus
Abstract
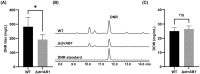
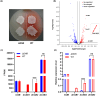
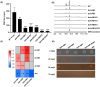
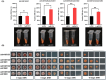
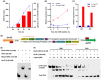
Streptomyces genus produces a large number of antibiotics, which are always synthesized by specific biosynthetic gene clusters (BGCs). To resist autotoxicity, transporters encoded by genes located within BGC occasionally pump antibiotic along with transporter encoded by gene located outside BGC. Daunorubicin is an anthracycline antibiotic biosynthesized by Streptomyces species, playing a crucial role in the treatment of leukaemia. In existing studies, only one two-component ATP-binding cassette (ABC) transporter, encoded by drrA1-drrB1 (abbreviated as drrAB1) and located within the daunorubicin BGC, has been proven to extrude daunorubicin. In this work, two other two-component ABC transporters, encoded by drrAB2 and drrAB3 and located outside the cluster, were found to play the complementary role in daunorubicin efflux in S. coeruleorubidus. Disruption of three drrABs resulted in a 94% decrease in daunorubicin production. Furthermore, drrAB2 is regulated by the TetR family regulator DrrR1, responding to the intracellular accumulation of daunorubicin and suggesting its role in stress response and self-resistance. Although the homologues of DrrAB1 are only found in three anthracycline BGCs, the homologues of DrrAB2 and DrrAB3 are spread in many Streptomyces strains which do not contain any known anthracycline BGC. This indicates that DrrAB2 and DrrAB3 may recognize and extrude a broader range of substrates besides daunorubicin, thus playing a more extensive role in cellular detoxification.
© 2024 The Author(s). Microbial Biotechnology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Aubel‐Sadron, G. & Londos‐Gagliardi, D. (1984) Daunorubicin and doxorubicin, anthracycline antibiotics, a physicochemical and biological review. Biochimie, 66, 333–352. - PubMed
-
- Bérdy, J. (2005) Bioactive microbial metabolites. Journal of Antibiotics (Tokyo), 58, 1–26. - PubMed
-
- Chen, C. , Chen, H. , Zhang, Y. , Thomas, H.R. , Frank, M.H. , He, Y. et al. (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13, 1194–1202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

