Inhibition of SARS-CoV-2 growth in the lungs of mice by a peptide-conjugated morpholino oligomer targeting viral RNA
- PMID: 39376996
- PMCID: PMC11456799
- DOI: 10.1016/j.omtn.2024.102331
Inhibition of SARS-CoV-2 growth in the lungs of mice by a peptide-conjugated morpholino oligomer targeting viral RNA
Abstract
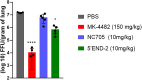
Further development of direct-acting antiviral agents against human SARS-CoV-2 infections remains a public health priority. Here, we report that an antisense peptide-conjugated morpholino oligomer (PPMO) named 5'END-2, targeting a highly conserved sequence in the 5' UTR of SARS-CoV-2 genomic RNA, potently suppressed SARS-CoV-2 growth in vitro and in vivo. In HeLa-ACE 2 cells, 5'END-2 produced IC50 values of between 40 nM and 1.15 μM in challenges using six genetically disparate strains of SARS-CoV-2, including JN.1. In vivo, using K18-hACE2 mice and the WA-1/2020 virus isolate, two doses of 5'END-2 at 10 mg/kg, administered intranasally on the day before and the day after infection, produced approximately 1.4 log10 virus titer reduction in lung tissue at 3 days post-infection. Under a similar dosing schedule, intratracheal administration of 1.0-2.0 mg/kg 5'END-2 produced over 3.5 log10 virus growth suppression in mouse lungs. Electrophoretic mobility shift assays characterized specific binding of 5'END-2 to its complementary target RNA. Furthermore, using reporter constructs containing SARS-CoV-2 5' UTR leader sequence, in an in-cell system, we observed that 5'END-2 could interfere with translation in a sequence-specific manner. The results demonstrate that direct pulmonary delivery of 5'END-2 PPMO is a promising antiviral strategy against SARS-CoV-2 infections and warrants further development.
Keywords: K-18-hACE2 mouse model; MT: Oligonucleotides: Therapies and Applications; PPMO; RNA virus; SARS-CoV-2; antisense; antiviral; intranasal delivery; intratracheal delivery; peptide-conjugated morpholino oligomer.
© 2024 The Author(s).
Conflict of interest statement
D.A.S. and H.M.M are listed as inventors on a US patent application, which includes a PPMO developed in this study, filed by Oregon State University. The H.M.M. laboratory has received research support from Radiation Control Technology, BriSight Biosciences, and Autoimmunity BioSolutions. The A.G.-S. laboratory has received research support from GSK, Pfizer, Senhwa Biosciences, Kenall Manufacturing, Blade Therapeutics, Avimex, Johnson & Johnson, Dynavax, 7 Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma, Applied Biological Laboratories, and Merck outside of the reported work. A.G.-S. has consulting agreements with the following companies involving cash and/or stock: Castlevax, Amovir, Vivaldi Biosciences, Contrafect, 7 Hills Pharma, Avimex, Pagoda, Accurius, Esperovax, Applied Biological Laboratories, Pharmamar, CureLab Oncology, CureLab Veterinary, Synairgen, Paratus, Pfizer, and Prosetta outside of the reported work. A.G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott and Astrazeneca. A.G.-S. is listed as an inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work. None of the support defined above is directly related to the research described in this paper.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

