USP26 Combats Age-Related Declines in Self-Renewal and Multipotent Differentiation of BMSC by Maintaining Mitochondrial Homeostasis
- PMID: 39377219
- PMCID: PMC11600297
- DOI: 10.1002/advs.202406428
USP26 Combats Age-Related Declines in Self-Renewal and Multipotent Differentiation of BMSC by Maintaining Mitochondrial Homeostasis
Abstract
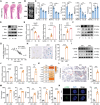
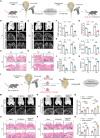
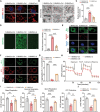
Age-related declines in self-renewal and multipotency of bone marrow mesenchymal stem cells (BMSCs) limit their applications in tissue engineering and clinical therapy. Thus, understanding the mechanisms behind BMSC senescence is crucial for maintaining the rejuvenation and multipotent differentiation capabilities of BMSCs. This study reveals that impaired USP26 expression in BMSCs leads to mitochondrial dysfunction, ultimately resulting in aging and age-related declines in the self-renewal and multipotency of BMSCs. Specifically, decreased USP26 expression results in decreased protein levels of Sirtuin 2 due to its ubiquitination degradation, which leads to mitochondrial dysfunction in BMSCs and ultimately resulting in aging and age-related declines in self-renewal and multilineage differentiation potentials. Additionally, decreased USP26 expression in aging BMSCs is a result of dampened hypoxia-inducible factor 1α (HIF-1α) expression. HIF-1α facilitates USP26 transcriptional expression by increasing USP26 promoter activity through binding to the -191 - -198 bp and -262 - -269 bp regions on the USP26 promoter. Therefore, the identification of USP26 as being correlated with aging and age-related declines in self-renewal and multipotency of BMSCs, along with understanding its expression and action mechanisms, suggests that USP26 represents a novel therapeutic target for combating aging and age-related declines in the self-renewal and multipotent differentiation of BMSCs.
Keywords: BMSC; USP26; aging; mitochondrial homeostasis; multipotency; self‐renewal.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- a) Hoang D. M., Pham P. T., Bach T. Q., Ngo A. T. L., Nguyen Q. T., Phan T. T. K., Nguyen G. H., Le P. T. T., Hoang V. T., Forsyth N. R., Heke M., Nguyen L. T., Signal Transduction Targeted Ther. 2022, 7, 272; - PMC - PubMed
- b) Uccelli A., Moretta L., Pistoia V., Nat. Rev. Immunol. 2008, 8, 726. - PubMed
-
- a) Deng P., Yuan Q., Cheng Y., Li J., Liu Z., Liu Y., Li Y., Su T., Wang J., Salvo M. E., Wang W., Fan G., Lyons K., Yu B., Wang C.‐Y., Cell Stem Cell 2021, 28, 1057; - PMC - PubMed
- b) Rauch A., Mandrup S., Nat. Rev. Mol. Cell Biol. 2021, 22, 465; - PubMed
- c) Zhang P., Dong J., Fan X., Yong J., Yang M., Liu Y., Zhang X., Lv L., Wen L., Qiao J., Tang F., Zhou Y., Signal Transduction Targeted Ther. 2023, 8, 126; - PMC - PubMed
- d) Liu F., Yuan Y., Bai L., Yuan L., Li L., Liu J., Chen Y., Lu Y., Cheng J., Zhang J., Redox Biol. 2021, 43, 101963. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
