Identification of RTS,S/AS01 vaccine-induced humoral biomarkers predictive of protection against controlled human malaria infection
- PMID: 39377226
- PMCID: PMC11466194
- DOI: 10.1172/jci.insight.178801
Identification of RTS,S/AS01 vaccine-induced humoral biomarkers predictive of protection against controlled human malaria infection
Abstract
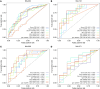
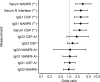
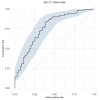
BACKGROUNDThe mechanism(s) responsible for the efficacy of WHO-recommended malaria vaccine RTS,S/AS01 are not completely understood. We previously identified RTS,S vaccine-induced Plasmodium falciparum circumsporozoite protein-specific (PfCSP-specific) antibody measures associated with protection from controlled human malaria infection (CHMI). Here, we tested the protection-predicting capability of these measures in independent CHMI studies.METHODSVaccine-induced total serum antibody (immunoglobulins, Igs) and subclass antibody (IgG1 and IgG3) responses were measured by biolayer interferometry and the binding antibody multiplex assay, respectively. Immune responses were compared between protected and nonprotected vaccinees using univariate and multivariate logistic regression.RESULTSBlinded prediction analysis showed that 5 antibody binding measures, including magnitude-avidity composite of serum Ig specific for PfCSP, major NANP repeats and N-terminal junction, and PfCSP- and NANP-specific IgG1 subclass magnitude, had good prediction accuracy (area under the receiver operating characteristic curves [ROC AUC] > 0.7) in at least 1 trial. Furthermore, univariate analysis showed a significant association between these antibody measures and protection (odds ratios 2.6-3.1). Multivariate modeling of combined data from 3 RTS,S CHMI trials identified the combination of IgG1 NANP binding magnitude plus serum NANP and N-junction Ig binding magnitude-avidity composite as the best predictor of protection (95% confidence interval for ROC AUC 0.693-0.834).CONCLUSIONThese results reinforce our previous findings and provide a tool for predicting protection in future trials.TRIAL REGISTRATIONClinicalTrials.gov NCT03162614, NCT03824236, NCT01366534, and NCT01857869.FUNDINGThis study was supported by Bill & Melinda Gates Foundation's Global Health-Discovery Collaboratory grants (INV-008612 and INV-043419) to GDT.
Keywords: Immunology; Malaria; Vaccines.
Figures


References
-
- World Health Organization. World malaria report 2023. World Health Organization; 2023. Accessed September 3, 2024. https://www.who.int/publications/i/item/9789240086173.
-
- World Health Organization. World malaria report 2022. World Health Organization; 2022. Accessed September 3, 2024. https://www.who.int/publications/i/item/9789240064898.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

