MYC-dependent upregulation of the de novo serine and glycine synthesis pathway is a targetable metabolic vulnerability in group 3 medulloblastoma
- PMID: 39377369
- PMCID: PMC11726242
- DOI: 10.1093/neuonc/noae179
MYC-dependent upregulation of the de novo serine and glycine synthesis pathway is a targetable metabolic vulnerability in group 3 medulloblastoma
Abstract
Background: Group 3 medulloblastoma (MBGRP3) represents around 25% of medulloblastomas and is strongly associated with c-MYC (MYC) amplification, which confers significantly worse patient survival. Although elevated MYC expression is a significant molecular feature in MBGRP3, direct targeting of MYC remains elusive, and alternative strategies are needed. The metabolic landscape of MYC-driven MBGRP3 is largely unexplored and may offer novel opportunities for therapies.
Methods: To study MYC-induced metabolic alterations in MBGRP3, we depleted MYC in isogenic cell-based model systems, followed by 1H high-resolution magic-angle spectroscopy (HRMAS) and stable isotope-resolved metabolomics, to assess changes in intracellular metabolites and pathway dynamics.
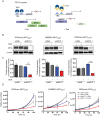
Results: Steady-state metabolic profiling revealed consistent MYC-dependent alterations in metabolites involved in one-carbon metabolism such as glycine. 13C-glucose tracing further revealed a reduction in glucose-derived serine and glycine (de novo synthesis) following MYC knockdown, which coincided with lower expression and activity of phosphoglycerate dehydrogenase (PHGDH), the rate-limiting enzyme in this pathway. Furthermore, MYC-overexpressing MBGRP3 cells were more vulnerable to pharmacological inhibition of PHGDH compared to those with low expression. Using in vivo tumor-bearing genetically engineered and xenograft mouse models, pharmacological inhibition of PHGDH increased survival, implicating the de novo serine/glycine synthesis pathway as a pro-survival mechanism sustaining tumor progression. Critically, in primary human medulloblastomas, increased PHGDH expression correlated strongly with both MYC amplification and poorer clinical outcomes.
Conclusions: Our findings support a MYC-induced dependency on the serine/glycine pathway in MBGRP3 that represents a novel therapeutic treatment strategy for this poor prognosis disease group.
Keywords: MYC; PHGDH; medulloblastoma; metabolism; serine.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Conflict of interest statement
O.D.K.M. is a co-founder, shareholder, and board member of Faeth Therapeutics and has contributed to patent application WO/2017/144877 by CRUK Cancer Research Technology. All other authors declare no competing interests.
Figures





References
-
- Massimino M, Biassoni V, Gandola L, et al.Childhood medulloblastoma. Crit Rev Oncol Hematol. 2016;105:35–51. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

