A role for BYN-1/bystin in cellular uptake and clearance of residual bodies in the Caenorhabditis elegans germline
- PMID: 39377446
- PMCID: PMC11488650
- DOI: 10.1242/dev.202694
A role for BYN-1/bystin in cellular uptake and clearance of residual bodies in the Caenorhabditis elegans germline
Abstract
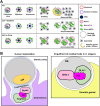
GLH/Vasa/DDX4 helicases are core germ-granule proteins that promote germline development and fertility. A yeast-two-hybrid screen using Caenorhabditis elegans GLH-1 as bait identified BYN-1, the homolog of human bystin/BYSL. In humans, bystin promotes cell adhesion and invasion in gliomas, and, with its binding partner trophinin, triggers embryonic implantation into the uterine wall. C. elegans embryos do not implant and lack a homolog of trophinin, but both trophinin and GLH-1 contain unique decapeptide phenylalanine-glycine (FG)-repeat domains. In germ cells, we find endogenous BYN-1 in the nucleolus, partitioned away from cytoplasmic germ granules. However, BYN-1 enters the cytoplasm during spermatogenesis to colocalize with GLH-1. Both proteins become deposited in residual bodies (RBs), which are then engulfed and cleared by the somatic gonad. We show that BYN-1 acts upstream of CED-1 to drive RB engulfment, and that removal of the FG-repeat domains from GLH-1 and GLH-2 can partially phenocopy byn-1 defects in RB clearance. These results point to an evolutionarily conserved pathway whereby cellular uptake is triggered by the cytoplasmic mobilization of bystin/BYN-1 to interact with proteins harboring FG-repeats.
Keywords: BYN-1; Bystin; FG-repeat; GLH-1; Residual bodies; Trophinin.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
-
- Arai, Y., Hosoda, F., Kobayashi, H., Arai, K., Hayashi, Y., Kamada, N., Kaneko, Y. and Ohki, M. (1997). The inv(11)(p15q22) chromosome translocation of de novo and therapy-related myeloid malignancies results in fusion of the nucleoporin gene, NUP98, with the putative RNA helicase gene, DDX10. Blood 89, 3936-3944. 10.1182/blood.V89.11.3936 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

