Epitranscriptomics and cervical cancer: the emerging role of m6A, m5C and m1A RNA modifications
- PMID: 39377535
- PMCID: PMC11488341
- DOI: 10.1017/erm.2024.20
Epitranscriptomics and cervical cancer: the emerging role of m6A, m5C and m1A RNA modifications
Abstract
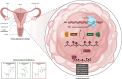
Cervical cancer (CC), one of the most prevalent and detrimental gynaecologic cancers, evolves through genetic and epigenetic alterations resulting in the promotion of oncogenic activity and dysfunction of tumour-suppressing mechanisms. Despite medical advancement, the prognosis for advanced-stage patients remains extremely low due to high recurrence rates and resistance to existing treatments. Thereby, the search for potential prognostic biomarkers is heightened to unravel new modalities of CC pathogenesis and to develop novel anti-cancer therapies. Epitranscriptomic modifications, reversible epigenetic RNA modifications, regulate various biological processes by deciding RNA fate to mediating RNA interactions. This narrative review provides insight into the cellular and molecular roles of endogenous RNA-editing proteins and their associated epitranscriptomic modifications, especially N6-methyladenosine (m6A), 5-methylcytosine (m5C) and N1-methyladenosine (m1A), in governing the development, progression and metastasis of CC. We discussed the in-depth epitranscriptomic mechanisms underlying the regulation of over 50 RNAs responsible for tumorigenesis, proliferation, migration, invasion, survival, autophagy, stemness, epithelial-mesenchymal transition, metabolism (glucose, lipid, glutamate and glutamine), resistance (drug and radiation), angiogenesis and recurrence of CC. Additionally, we provided a concise overview of the therapeutic potential of targeting the altered expression of endogenous RNA-editing proteins and aberrant deposition of RNA modifications on both coding and non-coding RNAs in CC.
Keywords: RNA modifications; RNA-editing proteins; anti-cancer therapies; cervical cancer; diagnostic biomarkers; epitranscriptomics; gene expression; m1A; m5C; m6A.
Conflict of interest statement
None.
Figures